Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mayra Alejandra Aguirre Garcia | + 2760 word(s) | 2760 | 2022-01-04 07:01:58 | | | |
| 2 | Jason Zhu | Meta information modification | 2760 | 2022-01-07 02:35:23 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Aguirre Garcia, M. Intestinal Organoids. Encyclopedia. Available online: https://encyclopedia.pub/entry/17836 (accessed on 07 February 2026).
Aguirre Garcia M. Intestinal Organoids. Encyclopedia. Available at: https://encyclopedia.pub/entry/17836. Accessed February 07, 2026.
Aguirre Garcia, Mayra. "Intestinal Organoids" Encyclopedia, https://encyclopedia.pub/entry/17836 (accessed February 07, 2026).
Aguirre Garcia, M. (2022, January 06). Intestinal Organoids. In Encyclopedia. https://encyclopedia.pub/entry/17836
Aguirre Garcia, Mayra. "Intestinal Organoids." Encyclopedia. Web. 06 January, 2022.
Copy Citation
Following advances in primary cell culture techniques, organoids have been developed. Such technological breakthroughs have opened a new path in the study of microbial infectious diseases, and thus opened onto new strategies to control foodborne hazards.
pathogenic mechanism
foodborne bacteria
organoids
enteroids
1. Introduction
Foodborne diseases (FBDs) are thought to be a major public health issue that contributes significantly to human morbidity and mortality around the world. The World Health Organization (WHO) estimates that almost one person in 10 falls ill from eating unsafe food every year [1]. Although the European region has the lowest burden in the world, the WHO calculated that more than 23 million people become sick annually because of FBDs [2]. Moreover, foodborne hazards of microbial origin raise a broad number of issues due to their economic burden. The European Food Safety Authority (EFSA) has estimated that the overall economic impact of human salmonellosis in Europe could be as high as EUR 3 billion annually [3]. In addition, antibiotic resistance and increasing food contamination as a consequence of environmental changes and dynamic methods of food production threaten to compound this problem further [4].
The surveillance of FBDs and our ability to tackle the knowledge gaps regarding host–pathogen–environment interactions need to be improved for the better prevention and control of microbial foodborne poisoning. Despite significant results from a large number of studies, their pathophysiology still appears to be poorly characterized, even less so where the pathogen can spread to distant organs and tissues through the blood stream and cause severe complications. One permanent challenge in this area of study is the lack of experimental models to address infection mechanisms and establish a clear picture of FBD biology.
2. Moving from Cell Lines to Intestinal Organoids
The oral route is the main entry site of FBPs, and the primary site of infection is the gastrointestinal tract [5]. They generally induce mild to severe enteritis, with widely known symptoms [6]. Because of this common pattern of infection, studies have been mostly focused on what occurs at the intestinal interface. The biology of these diseases remains less explored in other tissues [7], even though FBPs may occasionally spread deeply in the tissues and cause severe complications, permanent disability, and death [8][9][10].
From a historical perspective of model development and attempts to characterize bacterial FBP pathogenesis, concerns have emerged regarding animal models because bacterial intestinal pathogenesis varies considerably between humans and animals and the occurrence of symptoms in animals remains rare [11]. For example, Campylobacter jejuni and Salmonella enterica, both considered the main causes of bacterial FBDs worldwide, are mainly responsible for asymptomatic intestinal carriage in livestock [12]. In addition, national and international legislation and regulations restrict the use of animals in scientific procedures. The 3Rs principle (replacement, reduction, and refinement) aims to reduce the number of animals used in experimentation, which has led to the development of alternative methods [13]. In view of this, cell culture models of bacterial interaction with the epithelium have proved valuable for defining bacterium–host interactions [11].
The gold standard in intestinal modelling is based on immortalized cancer-derived cell lines, such as the enterocyte-like Caco-2 cell line. Numerous conclusions have been drawn from infected polarized or unpolarized cell monolayers (Figure 1a), even though it has been widely demonstrated over the last 50 years that these cell systems are outperformed [14]. As they consist of tumor-derived cells, they may not represent the native and healthy human intestine [15]. Several factors are likely to define intestinal homeostasis, and these vary considerably between cancer cell lines and the epithelial cells of native organs [16]. Structurally speaking, cell monolayers do not account for three-dimensional (3D) architecture and the complex cell population of the intestinal epithelium.
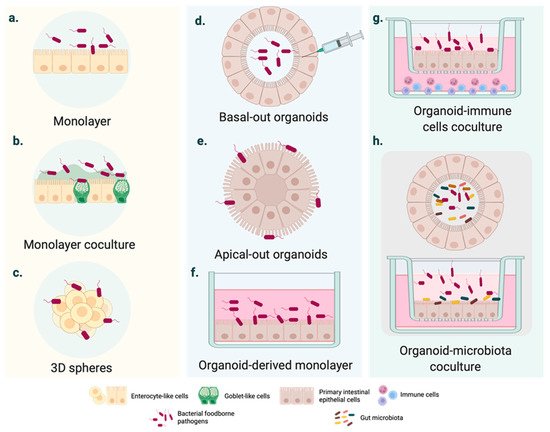
Figure 1. Cell culture systems mimicking intestinal FBD. (a–c) Intestinal FBD models derived from immortalized cells. (a) Polarized homogeneous cell monolayer typically based on immortalized cell lines with an enterocyte-like phenotype (e.g., Caco-2 cell monolayer). (b) Heterogeneous cell monolayer coculturing different cell lines to mimic essential intestinal features, such as the mucus- carrying intestinal tissue (e.g., Caco-2 and HT29 co-culture in vitro cell models). (c) 3D cell spheres developed from tumor-derived cell lines. (d–f) Intestinal organoid cultures generated from pluripotent stem cells (PSCs) or adult stem cells (AdSCs). (d) Basal-out organoid. The pathogen is generally injected inside the organoid. (e) Apical-out organoids might enhance the access of FBP with a high preference for the apical intestinal compartment. (f) Organoid-derived monolayers are D cell infection systems, such as the conventional immortalized cell cultures. (g–h) Coculture of intestinal organoids with immune cells and microbiota. More sophisticated organoid-based cultures, including intestinal epithelium–immune system and epithelium–microbiota interactions during infection.
In light of these disadvantages, cell coculture systems have been used to mirror the physiology of the human intestine more consistently. For instance, triple or cell coculture models (Figure 1b) have represented mucus-carrying intestinal tissue and basic elements of the innate immune system [17][18][19][20][21]. In parallel, the rotating wall vessel (RWV) facilitated the intestinal cell aggregation and growth in three dimensions (Figure 1c). Three-dimensional spheres resemble the native intestinal epithelium more accurately than monolayers derived from the same cell line [22]. The responses to bacterial pathogens also differ from those observed in 2D cell models [22][23].
Owing to the potential of organoids, the number of citations including the term “organoid” has rocketed in the last years. However, there does not seem to be a consensus on a general definition of organoids in the literature. In order to avoid misunderstandings, the recent definition suggested by Fujii and Sato was adopted in this review [24], i.e., ‘‘any heterotypic structures that can be reproducibly generated from single cells or cell clusters derived from somatic tissues or pluripotent stem cells, can self-assemble through cell–cell and cell–extracellular matrix (ECM) communications, and have some features of counterpart in vivo tissues’’ [24]. A further distinction is made according to the type of stem cell used to generate the organoids. While intestinal human organoids can be derived from pluripotent stem cells (PSCs) (including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs)) (Figure 2), adult stem cell (AdSC)-based organoids are initiated from self-renewing tissues, such as the gastrointestinal epithelium (see Figure 2) [25][26]. Two additional terms, enteroids and colonoids, are often used in the context of organoids to refer to the 3D models derived from intestinal and colon adult stem cells that only comprise epithelial cells (Figure 2) [27].
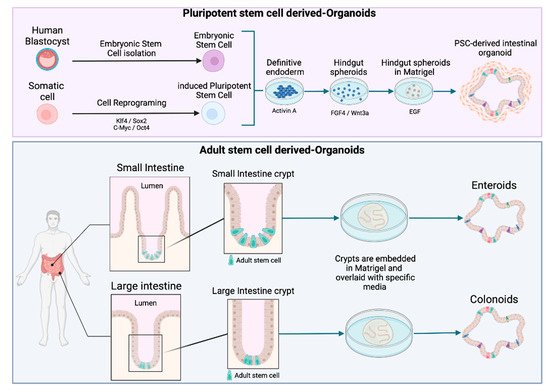
Figure 2. Schematic diagram of intestinal organoid, enteroid, and colonoid generation. Organoids can be derived from pluripotent stem cells (PSCs), including either induced pluripotent stem cells (iPSC) or embryonic stem cells (ESC). Enteroids and colonoids can be grown from the adult stem cells (AdSC) isolated from intestinal crypts.
Contrary to immortalized cancer-derived cell lines, intestinal organoids are characterized by the capacity to generate crypt-like domains with proliferative regions able to differentiate into all of the epithelial cell lineages.
To mimic the architectural and physiological properties of the in vivo small intestine, the models for foodborne diseases require differentiated crypt-villus structures. Intestinal crypts contain stem cells, which maintain the epithelial progenitor cells pool. Once generated, epithelial cells migrate toward the lumen, and differentiate and die at the tip of the villi. This process leads to a complete regeneration of the intestinal epithelium every 4–5 days [28]. Organoid culture is based on the capacity of the intestinal epithelial stem cells to perpetually divide and produce epithelial progenitor cells. The discovery of Lgr5 (Leucine-rich repeat-containing G protein-coupled receptor 5) has paved the way for culturing adult stem cells [29]. Lgr5+ intestinal stem cells cultured in 3D can undergo multi-lineage differentiation to ultimately form a “mini-gut”. In 2009, Sato et al. developed this long-term culture based on crucial signaling pathways, such as the Wnt/β-Catenin pathway and the EGF/EGF receptor (EGFR) with ECM-supported culture [30]. The resulting organoid culture system has been successfully applied to culture other epithelial organs, including stomach, pancreas, colon, and liver organoids [14].
Organoids have been mainly used for the study of cancer and genetic disorders as well as host cell–microorganism interactions [31]. In the organoid–pathogen coculture, several constraints in the mimicking of viral and human host-specific infections have been overcome. Alternatively, organoids generated from genetically modified pluripotent stem cells or from patients harboring mutations of clinical interest have opened a new window onto human infection diseases [32]. Furthermore, these practical and reproducible in vitro models of infection lead to the exploration of additional host–microbe dynamics, e.g., in disseminated infections [7][33][34].
Intestinal organoids usually form structures with budded and branched shapes [35], encapsulating the apical surface and the lumen (Figure 1d) [36]. This makes pathogen delivery inside the organoid interior more challenging from a technical point of view. Even though several studies have employed microinjection (Figure 1e), this is a tedious technique and observations can be disturbed by cellular material accumulating within the luminal side; moreover, cellular material may damage the organoid epithelium [36].
In 2019, Co et al. developed a culture system where organoids could precisely adopt polarity-specific parameters inspired by previous studies of polarity reversal in Madin–Darby canine kidney (MDCK) spheroids [36][37]. The resulting method provided a cell apparatus with an apical-out surface that promoted pathogen inclusion, especially of microbes with a marked preference for interacting with the apical intestinal compartment [36].
Although the study of intestinal epithelial cell (IEC)–pathogen interactions is time and cell consuming [36], most studies have used organoid-derived monolayers on insert/filter membranes (Figure 1f). Two-dimensional cell systems, as with other conventional transmembrane models, provide experimental access to the apical or the basolateral surface [38]. Similarly, monolayers of somatic cells allow adding other nearby intestinal cells to transformed cell lines in coculture to analyze the cellular crosstalk associated with the response to infection (Figure 1g) [39][40]. Although these complex cell systems are still in their infancy, advances have been made in modeling the intestinal microenvironment systems containing macrophages and T-cells (Figure 1g) [39][41] or microbiota (Figure 1h). On a wider scale, hybrid cell cultures could provide insights into the tissue inflammation and carcinogenesis significantly associated with intestinal infections.
3. Using Organoids to Explore the Cell and Tissue Tropism of FBPs
Regarding the infection capacity of FBPs, plausible discrepancies can be observed between homogenous cell monolayers and organoids that retain most of the intestinal cell composition and somatic signatures. Early works have shown that bacteria can cause the loss of a tissue’s structural integrity in intestinal organoids. Unsurprisingly, a growing body of evidence has assessed this common and fundamental issue. Antibiotic-protection assays coupled to confocal imaging to evaluate changes of the actin network have showed that Salmonella-, enterohemorrhagic Escherichia coli (EHEC)-, Listeria monocytogenes-, or Shigella-infected organoids showed intracellular pathogen carriage and damage of intestinal tissue in vitro [36][42][43][44].
Upon reaching the intestinal epithelium, some pathogens exhibit a higher affinity for regional intestinal segments [45]. Enteroids derived from cells from an anatomical region of the intestine could be a potential starting point for reliably studying segment-specific colonization on an in vitro device, an achievement never attained in whole animal models [46]. VanDussen et al. inoculated various strains of pathogenic E. coli to the apical surface of a cell monolayer generated from the dissociation of human intestinal biopsies [38]. E. coli EPEC strains preferentially adhered to ileal epithelial cells, whereas E. coli EAggEC and EHEC strains instead adhered to rectal epithelial cells. In et al. noted a remarkable difference between the number of EHEC bacteria associated with the apical surface in organoids representing colon and jejunum environments [43]. The authors indicated that the preference of EHEC for these colonoids could be related to the colon-specific differentiation [43]. Each E. coli pathotype usually possesses distinct virulence mechanisms to disrupt the host intestinal epithelium. Adherence patterns are one of the key signs generally accepted among E. coli pathovars [47]. Rajan et al. mimicked bacterial adhesion using enteroids made from crypts isolated from tissues from four different gut segments. Histopathological comparisons of infected enteroids suggested that E. coli EAggEC aggregated in several ways, including those patterns observed in classic in vitro models and new ones, with a remarkable dependency on donor and intestinal segment tropism [48].
Unlike EHEC, Shigella flexneri can invade enteroids from the duodenum, ileum, and colon in the same manner [49]. However, these findings substantially contrast with the in vivo shigellosis biology that describes a specificity of Shigella to the rectal and colonic mucosae [50]. Thus, other elements of the intestinal microenvironment, such as vasculature, the enteric nervous system, or the resident microbiota contributing human colon infection, were not taken into account with the previous enteroid study [49].
Several studies have showed the preferential attachment of FBP on the apical surface of immortalized cell lines [11][20][51][52][53]. However, some works have investigated the ability of enterocytes to internalize bacteria for transcellular translocation from the basolateral to the apical compartment. To address this issue, Co et al. developed a reversed polarity apical-out human enteroid model [36]. Thanks to this novel cell culture platform, they were able to compare the binding patterns of S. enterica Typhimurium and L. monocytogenes. Salmonella predominantly invade apical-out enteroids and induce cytoskeletal rearrangement, as described using cancer derived monolayers [54]. Conversely, the Gram-positive L. monocytogenes adhered more to the basal-out enteroids. When the author used mixed polarity enteroids, whose polarity had been partially reversed and contained both basal-out and apical-out surfaces, both pathogens preferentially invaded the apical side [36]. Apical-out human enteroids seem to be relevant and accessible models because they highlight the importance of cell polarity to visualize the mechanism of pathogen exit from the epithelium to promote shedding and dissemination. This is particularly true for pathogens that use basolateral receptors for invasion, such as L. monocytogenes or S. flexneri.
Organoids can be used to model the complex multicellular environment of the intestine. Experimental workflows now finely sum up the interactions of pathogens with highly specialized epithelia cells (i.e., mucus-producing cells, Paneth cells, and microfold (M) cells). This could overcome the limitations of the in vitro cell lines that commonly represent enterocytes [46].
The thick mucus layer is a key component of the physical barrier that protects the gut epithelium from the potential pathogens present in the luminal environment [55]. Transcript-based comparisons using organoids have showed changes in the expression signature of mucin Muc2, the major structural component of the intestinal mucus. A study based on fully differentiated enteroids infected with S. flexneri indicated the transcriptional upregulation of Muc2 after apical or basolateral bacterial infection [56]. Similar Muc2 transcript profiles were observed using the goblet-like cells HT29-MTX infected with S. flexneri [56]. While non-motile bacteria, such as Shigella, increased the level of Muc2, EHEC exposure to human colonoids reduce the thickness of the Muc2-positive mucus layer in less than 6 h [43].
The follicle-associated epithelium (FAE) is characterized by the presence of M cells, which constitute a niche for bacteria with an intracellular lifestyle because they naturally internalize foreign particles. M cells are exploited by many different pathogens, including S. flexneri [57], L. monocytogenes [58], and S. enterica Typhimurium [59], as a passage through the intestinal barrier to deeper host tissues [60]. S. enterica Typhimurium-infected enteroids derived from human small intestinal crypts confirmed that bacteria could rapidly trigger a transition from FAE enterocytes into M cells via an epithelial-mesenchymal transition (EMT) [61]. Similar findings were reported using cocultures of Caco-2 and Raji-B cells [62]. Stimulation with receptor activator of NF-κB Ligand (RANKL) and tumor necrosis factor alpha (TNF-α) was used to induce M cell differentiation in enteroids [63]. The resulting 3D intestinal in vitro device was used to study S. flexneri transcytosis via M cells [56]. The authors confirmed the presence of M cells using glycoprotein 2 immunostaining. S. flexneri invaded M cell-containing enteroids more often than it invaded non-stimulated enteroids [56].
FBDs are usually self-limiting and of short duration. Some FBD cases, however, can lead to long-lasting disability. A range of human tissues are currently expandable as organoids, but only a few applications are currently used to explore the interactions of FBPs with tissues or cells once the pathogen has colonized the deeper tissues. Organoids have been used to understand the molecular mechanisms behind the epidemiological association between chronic infection with Salmonella enterica and gallbladder carcinoma (GBC) in humans. Scanu et al. developed a murine gallbladder organoid (GBO) genetically predisposed to resemble the analogous TP53 inactivation in GBC patients. Infected murine cells formed organoids in growth factor-free medium. In addition, they presented polarity loss and large irregular nuclei. These observations indicate a cell transformation driven by Salmonella infection [64]. More recent evidence reveals that the human restricted pathogenic serovar Paratyphi A induced DNA damage in human GBO [7]. A detailed analysis of longer-term infected organoids reveals that bacteria could drive the termination of cell replication via the downregulation of the transcriptional programs related to each cell cycle phase (G1/S, S, G2, and G2/M) [7]. Therefore, these studies showed not only a clear Salmonella tropism of gallbladder tissue, but also the underlying pathways of the connection between S. enterica and cancer.
References
- WHO. WHO Estimates of the Global Burden of Foodborne Diseases; World Health Organization: Geneva, Switzerland, 2015; p. 254.
- WHO. The Burden of Foodborne Diseases in the WHO European Region; World Health Organization: Copenhagen, Danemark, 2017.
- EFSA. EFSA Explains Zoonotic Diseases: Salmonella. Available online: https://data.europa.eu/doi/10.2805/61217 (accessed on 10 November 2021).
- Newell, D.G.; Koopmans, M.; Verhoef, L.; Duizer, E.; Aidara-Kane, A.; Sprong, H.; Opsteegh, M.; Langelaar, M.; Jacobson, A.; Scheutz, F.; et al. Food-borne diseases—The challenges of 20years ago still persist while new ones continue to emerge. Int. J. Food Microbiol. 2010, 139, S3–S15.
- Drolia, R.; Bhunia, A.K. Crossing the intestinal barrier via Listeria adhesion protein and Internalin A. Trends Microbiol. 2019, 27, 408–425.
- Abebe, E.; Gugsa, G.; Ahmed, M. Review on major food-borne zoonotic bacterial pathogens. J. Trop. Med. 2020, 2020, 4674235.
- Sepe, L.P.; Hartl, K.; Iftekhar, A.; Berger, H.; Kumar, N.; Goosmann, C.; Chopra, S.; Schmidt, S.C.; Gurumurthy, R.K.; Meyer, T.F.; et al. Genotoxic effect of Salmonella Paratyphi A infection on human primary gallbladder cells. mBio 2020, 11, e01911-20.
- Mayer, C.L.; Leibowitz, C.S.; Kurosawa, S.; Stearns-Kurosawa, D.J. Shiga toxins and the pathophysiology of hemolytic uremic syndrome in humans and animals. Toxins 2012, 4, 1261–1287.
- McLauchlin, J.; Mitchell, R.T.; Smerdon, W.J.; Jewell, K. Listeria monocytogenes and listeriosis: A review of hazard characterisation for use in microbiological risk assessment of foods. Int. J. Food Microbiol. 2004, 92, 15–33.
- Rees, J.H.; Soudain, S.E.; Gregson, N.A.; Hughes, R.A. Campylobacter jejuni infection and Guillain-Barre syndrome. N. Engl. J. Med. 1995, 333, 1374–1379.
- Haddad, N.; Marce, C.; Magras, C.; Cappelier, J.M. An overview of methods used to clarify pathogenesis mechanisms of Campylobacter jejuni. J. Food Prot. 2010, 73, 786–802.
- EFSA; ECDC. The European Union one health 2018 zoonoses report. EFSA J. 2019, 17, e05926.
- Eskes, C. The usefulness of integrated strategy approaches in replacing animal experimentation. Ann. Ist. Super Sanita. 2019, 55, 400–404.
- Simian, M.; Bissell, M.J. Organoids: A historical perspective of thinking in three dimensions. J. Cell Biol. 2017, 216, 31–40.
- Ross, D.T.; Scherf, U.; Eisen, M.B.; Perou, C.M.; Rees, C.; Spellman, P.; Iyer, V.; Jeffrey, S.S.; Van de Rijn, M.; Waltham, M.; et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat. Genet. 2000, 24, 227–235.
- Langerholc, T.; Maragkoudakis, P.A.; Wollgast, J.; Gradisnik, L.; Cencic, A. Novel and established intestinal cell line models–An indispensable tool in food science and nutrition. Trends Food Sci. Technol. 2011, 22, S11–S20.
- Ahmad, I.; Lamprokostopoulou, A.; Le Guyon, S.; Streck, E.; Barthel, M.; Peters, V.; Hardt, W.-D.; Römling, U. Complex c-di-GMP signaling networks mediate transition between virulence properties and biofilm formation in Salmonella enterica serovar Typhimurium. PLoS ONE 2011, 6, e28351.
- Bahrami, B.; Macfarlane, S.; Macfarlane, G.T. Induction of cytokine formation by human intestinal bacteria in gut epithelial cell lines. J. Appl. Microbiol. 2011, 110, 353–363.
- Martinez-Argudo, I.; Jepson, M.A. Salmonella translocates across an in vitro M cell model independently of SPI-1 and SPI-2. Microbiology 2008, 154, 3887–3894.
- Rodrigues, R.C.; Pocheron, A.-L.; Cappelier, J.-M.; Tresse, O.; Haddad, N. An adapted in vitro assay to assess Campylobacter jejuni interaction with intestinal epithelial cells: Taking into stimulation with TNFα. J. Microbiol. Met. 2018, 149, 67–72.
- Zamora, C.Y.; Ward, E.M.; Kester, J.C.; Kelly Chen, W.L.; Velazquez, J.G.; Griffith, L.G.; Imperiali, B. Application of a gut-immune co-culture system for the study of N-glycan-dependent host-pathogen interactions of Campylobacter jejuni. Glycobiology 2020, 30, 374–381.
- Nickerson, C.A.; Goodwin, T.J.; Terlonge, J.; Ott, C.M.; Buchanan, K.L.; Uicker, W.C.; Emami, K.; LeBlanc, C.L.; Ramamurthy, R.; Clarke, M.S.; et al. Three-dimensional tissue assemblies: Novel models for the study of Salmonella enterica serovar Typhimurium pathogenesis. Infect. Immun. 2001, 69, 7106–7120.
- Höner zu Bentrup, K.; Ramamurthy, R.; Ott, C.M.; Emami, K.; Nelman-Gonzalez, M.; Wilson, J.W.; Richter, E.G.; Goodwin, T.J.; Alexander, J.S.; Pierson, D.L.; et al. Three-dimensional organotypic models of human colonic epithelium to study the early stages of enteric salmonellosis. Microb. Infect. 2006, 8, 1813–1825.
- Fujii, M.; Sato, T. Somatic cell-derived organoids as prototypes of human epithelial tissues and diseases. Nat. Mater. 2020, 20, 156–169.
- Clevers, H. Modeling development and disease with organoids. Cell 2016, 165, 1586–1597.
- Flatres, C.; Loffet, E.; Neunlist, M.; Mahe, M.M. From human pluripotent stem cells to custom-made intestinal organoids. Med. Sci. 2019, 35, 549–555.
- Mahe, M.M.; Sundaram, N.; Watson, C.L.; Shroyer, N.F.; Helmrath, M.A. Establishment of human epithelial enteroids and colonoids from whole tissue and biopsy. J. Vis. Exp. 2015, 97, e52483.
- Barker, N. Adult intestinal stem cells: Critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 2014, 15, 19–33.
- Barker, N.; van Es, J.H.; Kuipers, J.; Kujala, P.; van den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007, 449, 1003–1007.
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265.
- Dutta, D.; Clevers, H. Organoid culture systems to study host–pathogen interactions. Curr. Op. Immunol. 2017, 48, 15–22.
- Forbester, J.L.; Lees, E.A.; Goulding, D.; Forrest, S.; Yeung, A.; Speak, A.; Clare, S.; Coomber, E.L.; Mukhopadhyay, S.; Kraiczy, J.; et al. Interleukin-22 promotes phagolysosomal fusion to induce protection against Salmonella enterica Typhimurium in human epithelial cells. Proc. Natl. Acad. Sci. USA 2018, 115, 10118–10123.
- DesRochers, T.M.; Kimmerling, E.P.; Jandhyala, D.M.; El-Jouni, W.; Zhou, J.; Thorpe, C.M.; Leong, J.M.; Kaplan, D.L. Effects of Shiga toxin type 2 on a bioengineered three-dimensional model of human renal tissue. Infect. Immun. 2015, 83, 28–38.
- Marquez, L.B.; Araoz, A.; Repetto, H.A.; Ibarra, F.R.; Silberstein, C. Effects of shiga toxin 2 on cellular regeneration mechanisms in primary and three-dimensional cultures of human renal tubular epithelial cells. Microb. Pathog. 2016, 99, 87–94.
- Rozman, J.; Krajnc, M.; Ziherl, P. Collective cell mechanics of epithelial shells with organoid-like morphologies. Nat. Commun. 2020, 11, 3805.
- Co, J.Y.; Margalef-Català, M.; Li, X.; Mah, A.T.; Kuo, C.J.; Monack, D.M.; Amieva, M.R. Controlling epithelial polarity: A human enteroid model for host-pathogen interactions. Cell Rep. 2019, 26, 2509–2520.e4.
- Wang, A.Z.; Ojakian, G.K.; Nelson, W.J. Steps in the morphogenesis of a polarized epithelium. II. Disassembly and assembly of plasma membrane domains during reversal of epithelial cell polarity in multicellular epithelial (MDCK) cysts. J. Cell Sci. 1990, 95 Pt 1, 153–165.
- VanDussen, K.L.; Marinshaw, J.M.; Shaikh, N.; Miyoshi, H.; Moon, C.; Tarr, P.I.; Ciorba, M.A.; Stappenbeck, T.d.S. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 2015, 64, 911–920.
- Noel, G.; Baetz, N.W.; Staab, J.F.; Donowitz, M.; Kovbasnjuk, O.; Pasetti, M.F.; Zachos, N.C. A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci. Rep. 2017, 7, 45270.
- Yuki, K.; Cheng, N.; Nakano, M.; Kuo, C.J. Organoid models of tumor immunology. Trends Immunol. 2020, 41, 652–664.
- Rogoz, A.; Reis, B.S.; Karssemeijer, R.A.; Mucida, D. A 3-D enteroid-based model to study T-cell and epithelial cell interaction. J. Immunol. Methods 2015, 421, 89–95.
- Holly, M.K.; Han, X.; Zhao, E.J.; Crowley, S.M.; Allaire, J.M.; Knodler, L.A.; Vallance, B.A.; Smith, J.G. Salmonella enterica infection of murine and human enteroid-derived monolayers elicits differential activation of epithelial-intrinsic inflammasomes. Infect. Immun. 2020, 88, e00017-20.
- In, J.; Foulke-Abel, J.; Zachos, N.C.; Hansen, A.M.; Kaper, J.B.; Bernstein, H.D.; Halushka, M.; Blutt, S.; Estes, M.K.; Donowitz, M.; et al. Enterohemorrhagic Escherichia coli reduce mucus and intermicrovillar bridges in human stem cell-derived colonoids. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 48–62.e3.
- Karve, S.S.; Pradhan, S.; Ward, D.V.; Weiss, A.A. Intestinal organoids model human responses to infection by commensal and Shiga toxin producing Escherichia coli. PLoS ONE 2017, 12, e0178966.
- McCall, L.I.; Siqueira-Neto, J.L.; McKerrow, J.H. Location, location, location: Five facts about tissue tropism and pathogenesis. PLoS Pathog. 2016, 12, e1005519.
- In, J.G.; Foulke-Abel, J.; Estes, M.K.; Zachos, N.C.; Kovbasnjuk, O.; Donowitz, M. Human mini-guts: New insights into intestinal physiology and host–pathogen interactions. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 633–642.
- Harrington, S.M.; Dudley, E.G.; Nataro, J.P. Pathogenesis of enteroaggregative Escherichia coli infection. FEMS Microbiol. Lett. 2006, 254, 12–18.
- Rajan, A.; Vela, L.; Zeng, X.-L.; Yu, X.; Shroyer, N.F.; Blutt, S.E.; Poole, N.M.; Carlin, L.G.; Nataro, J.P.; Estes, M.K.; et al. Novel segment- and host-specific patterns of enteroaggregative Escherichia coli adherence to human intestinal enteroids. mBio 2018, 9, e02419-17.
- Koestler, B.J.; Ward, C.M.; Fisher, C.R.; Rajan, A.; Maresso, A.W.; Payne, S.M. Human intestinal enteroids as a model system of Shigella pathogenesis. Infect. Immun. 2019, 87, e00733-18.
- Sansonetti, P.J. Rupture, invasion and inflammatory destruction of the intestinal barrier by Shigella, making sense of prokaryote-eukaryote cross-talks. FEMS Microbiol. Rev. 2001, 25, 3–14.
- Hamon, M.; Bierne, H.; Cossart, P. Listeria monocytogenes: A multifaceted model. Nat. Rev. Microbiol. 2006, 4, 423–434.
- Knodler, L.A.; Crowley, S.M.; Sham, H.P.; Yang, H.; Wrande, M.; Ma, C.; Ernst, R.K.; Steele-Mortimer, O.; Celli, J.; Vallance, B.A. Noncanonical inflammasome activation of Caspase-4/Caspase-11 mediates epithelial defenses against enteric bacterial pathogens. Cell Host Microb. 2014, 16, 249–256.
- Pentecost, M.; Kumaran, J.; Ghosh, P.; Amieva, M.R. Listeria monocytogenes Internalin B activates junctional endocytosis to accelerate intestinal invasion. PLoS Pathog. 2010, 6, e1000900.
- Galán, J.E. Salmonella interactions with host cells: Type III secretion at work. Annu. Rev. Cell Dev. Biol. 2001, 17, 53–86.
- Johansson, M.E.; Phillipson, M.; Petersson, J.; Velcich, A.; Holm, L.; Hansson, G.C. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 15064–15069.
- Ranganathan, S.; Doucet, M.; Grassel, C.L.; Delaine-Elias, B.; Zachos, N.C.; Barry, E.M. Evaluating Shigella flexneri pathogenesis in the human enteroid model. Infect. Immun. 2019, 87, e00740-18.
- Rey, C.; Chang, Y.-Y.; Latour-Lambert, P.; Varet, H.; Proux, C.; Legendre, R.; Coppée, J.-Y.; Enninga, J. Transcytosis subversion by M cell-to-enterocyte spread promotes Shigella flexneri and Listeria monocytogenes intracellular bacterial dissemination. PLoS Pathog. 2020, 16, e1008446.
- Chiba, S.; Nagai, T.; Hayashi, T.; Baba, Y.; Nagai, S.; Koyasu, S. Listerial invasion protein internalin B promotes entry into ileal Peyer’s patches in vivo. Microbiol. Immunol. 2011, 55, 123–129.
- Jones, B.D.; Ghori, N.; Falkow, S. Salmonella Typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer’s patches. J. Exp. Med. 1994, 180, 15–23.
- Mabbott, N.A.; Donaldson, D.S.; Ohno, H.; Williams, I.R.; Mahajan, A. Microfold (M) cells: Important immunosurveillance posts in the intestinal epithelium. Mucosal Immunol. 2013, 6, 666–677.
- Rouch, J.D.; Scott, A.; Lei, N.Y.; Solorzano-Vargas, R.S.; Wang, J.; Hanson, E.M.; Kobayashi, M.; Lewis, M.; Stelzner, M.G.; Dunn, J.C.Y.; et al. Development of functional Microfold (M) cells from intestinal stem cells in primary human enteroids. PLoS ONE 2016, 11, e0148216.
- Tahoun, A.; Mahajan, S.; Paxton, E.; Malterer, G.; Donaldson, D.S.; Wang, D.; Tan, A.; Gillespie, T.L.; O’Shea, M.; Roe, A.J.; et al. Salmonella transforms follicle-associated epithelial cells into M cells to promote intestinal invasion. Cell Host Microb. 2012, 12, 645–656.
- Wood, M.B.; Rios, D.; Williams, I.R. TNF-α augments RANKL-dependent intestinal M cell differentiation in enteroid cultures. Am. J. Physiol. Cell. Physiol. 2016, 311, C498–C507.
- Scanu, T.; Spaapen, R.M.; Bakker, J.M.; Pratap, C.B.; Wu, L.-E.; Hofland, I.; Broeks, A.; Shukla, V.K.; Kumar, M.; Janssen, H.; et al. Salmonella manipulation of host signaling pathways provokes cellular transformation associated with gallbladder carcinoma. Cell Host Microb. 2015, 17, 763–774.
More
Information
Subjects:
Agriculture, Dairy & Animal Science
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.4K
Revisions:
2 times
(View History)
Update Date:
07 Jan 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




