Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Kristine Y. DeLeon-Pennell | + 1691 word(s) | 1691 | 2021-12-28 07:13:28 | | | |
| 2 | Dean Liu | Meta information modification | 1691 | 2021-12-30 01:39:04 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Deleon-Pennell, K.Y. Technologies Used to Discover Immune Cell Heterogeneity. Encyclopedia. Available online: https://encyclopedia.pub/entry/17629 (accessed on 02 March 2026).
Deleon-Pennell KY. Technologies Used to Discover Immune Cell Heterogeneity. Encyclopedia. Available at: https://encyclopedia.pub/entry/17629. Accessed March 02, 2026.
Deleon-Pennell, Kristine Y.. "Technologies Used to Discover Immune Cell Heterogeneity" Encyclopedia, https://encyclopedia.pub/entry/17629 (accessed March 02, 2026).
Deleon-Pennell, K.Y. (2021, December 29). Technologies Used to Discover Immune Cell Heterogeneity. In Encyclopedia. https://encyclopedia.pub/entry/17629
Deleon-Pennell, Kristine Y.. "Technologies Used to Discover Immune Cell Heterogeneity." Encyclopedia. Web. 29 December, 2021.
Copy Citation
During homeostasis, immune cells perform daily housekeeping functions to maintain heart health by acting as sentinels for tissue damage and foreign particles.
cardiovascular disease
myocardial infarction
pressure overload
1. Introduction
In response to injury after myocardial infarction (MI), there is an increase in inflammation within the heart due to the influx of immune cells. Some of the most common immune cells recruited to the heart are macrophages, dendritic cells, monocytes, neutrophils, and T-cells [1][2]. Inflammation after MI must reside within a Goldilocks zone to have a beneficial effect. The Goldilocks zone is defined as the balance of immune cells necessary for efficient cardiac healing. While some inflammation is essential for clearance of tissue damage, excessive or prolonged inflammation can be detrimental as it may lead to increased cardiac rupture from disproportionate collagen degradation, infarct expansion via phagocytosis of healthy cardiomyocytes, and/or increased dilation of the left ventricle (LV; Figure 1A) [2][3]. Knowing the stimuli that facilitates recruitment and phenotypic changes within cell types can help uncover regulators of beneficial versus adverse cardiac remodeling [4].
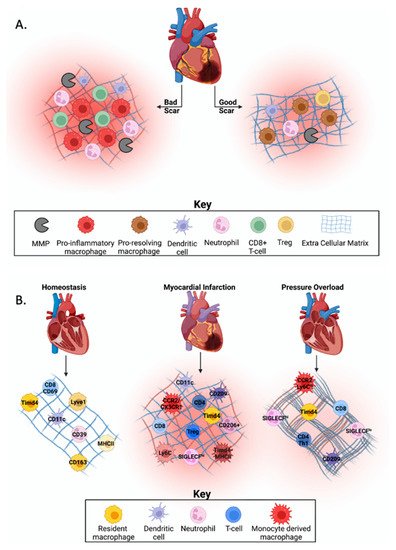
Figure 1. Immune cell heterogeneity in cardiovascular homeostasis and disease.
Heart failure with preserved ejection fraction (HFpEF) is a diagnosis frequently associated with conditions that give rise to increased pressure overload (PO) on the heart including hypertension and aortic valve disease. A hallmark of HFpEF is an increase in myocardial stiffness contributed by both changes in cardiac myocyte sarcomeres and increases in interstitial collagen content [5]. More recently, there has been a shift in the focus to include the role of immune cells in regulating HFpEF pathology. While the role of immune cells post-MI is better understood, fewer studies have focused on how these immune cells regulate each other in the heart before and after PO.
2. Technologies Used to Discover Immune Cell Heterogeneity
Studying immune cell heterogeneity in the heart before and after injury is quite complex. Many immune cells express the same cell surface receptors and proteins making it tough to distinguish certain immune cell groups from others. For example, CD44 is expressed on both T-cells and macrophages. CD44 has many roles in proliferation, migration, and lymphocyte activation [6]. Similarly, CD8, which is a key marker for cytotoxic T-cells, is also expressed by dendritic cells [7]. Because of the overlap in immune cell markers to classify cells into the correct subgroups, multiple markers should be used to positively capture the desired target cell population. Recent advances in technology used to study cell heterogeneity have made it easier to isolate and characterize immune cell populations with increased precision and accuracy. Below we discuss technological advancements for understanding cellular heterogeneity in models of cardiac disease and summarize the advantages and limitations of these techniques. Understanding the limitations of each method and how different techniques complement one another is vital when designing experiments to study the role of the immune system in the heart. Nonetheless, great promise is seen in utilizing new techniques for the future of cardiac research and eventually, the development of novel therapies.
2.1. Flow Cytometry
Flow cytometry is used to detect and measure physical and chemical properties of isolated cells. Utilizing antibodies for specific immune cell classification and phenotypes can define the immune cell composition within tissue, serum, plasma, etc. Flow cytometry can also be used to phenotype cells by measuring cellular proliferation, and viability, in addition to intracellular cytokine and signaling proteins, and cell cycle stages [8].
Fluorescent activated cell sorting (FACS) is a way to enrich for a cell population of interest from a sample by using flow cytometry technology. The cells of a pre-selected population can be isolated and sorted rapidly with high purity; some reports indicate a purity of up to 99% [8][9]. Sorting is performed by giving charges to droplets containing single cells. Single cells are then detected by an electric field and sorted into collection tubes according to their charge [8]. After sorting, enriched cells are often sent for sequencing, biochemical analysis, or grown in culture.
Strengths and Limitations
Flow cytometry and FACS techniques are fairly easy to use, highly translational, and affordable [8]. Because flow cytometry data removes debris and dead cells, the accuracy is much better than more traditional techniques such as immunofluorescence. One limitation of flow cytometry and FACS is that these techniques are dependent on antibodies or stains to characterize the cells. In addition, because the number of detectors/filters available to use are limited, some immune cell populations will be difficult to analyze. However, recent advances have greatly increased the number of fluorescently tagged antibodies available for flow cytometry. Available fluorophores with distinct excitation and emission spectra along with multicolor flow cytometers have enhanced the ability to generate multidimensional expression data but choosing fluorochromes with minimal spectral overlap to fully characterize the cell sub-populations remains tricky [8]. Online resources such as Fluorofinder, Spectrum Viewer, Fluorescence Spectra Viewer, or the Spectra Analyzer can help design antibody panels and assess the degree of spectral overlap and potential spillover. Given the lack of reagents for most animal models, these advanced techniques have mainly benefitted the analysis of human and mouse cell samples. In addition, because samples are in suspension, spatial and cellular interaction data are not easily attainable. Overall, flow cytometry and FACS techniques have contributed considerably to identifying cellular phenotypes of immune cell populations in the heart.
2.2. Mass Spectrometry
Mass spectrometry and its applications are being used more readily at the bench and clinic. Matrix-assisted laser desorption/ionization- mass spectrometry (MALDI-MS) is an extremely sensitive technique used for high throughput proteomic assessment of tissue and cellular samples which uses the intensity of the spectrum to determine the abundance of the compound [10][11]. MALDI imaging mass spectrometry (MALDI-IMS) is a specialized proteomic method that can identify the molecular composition and abundance while also giving spatial distribution of proteins in a tissue section [12]. MALDI-IMS measures the mass spectra at specific spatial points on the sample to provide a hyperspectral image with a corresponding mass spectrum at each measured pixel, which then corresponds to specific compounds and their location in the sample [11]. MALDI-IMS is useful to visualize changes in the microenvironment including alterations in chemokines, ECM, and growth factors, that might directly influence cellular recruitment and activation, leading to impaired states of wound healing.
Cytometry by time of flight (CyTOF), is a powerful single-cell proteomic analysis technique which utilizes rare metal isotopes instead of fluorophores as antibody tags. The application of CyTOF is similar to flow cytometry and often used to quantify labeled intracellular proteins or cell surface targets. Because CyTOF uses metal isotopes instead of fluorophores, a larger number of markers can be assessed while remaining high-dimensional and unbiased [13].
Strengths and Limitations
Mass spectrometry detection is flexible as it can detect proteins, lipids, cell metabolism, and drug metabolites on one platform making this method applicable to many fields [14]. The advantages of using proteomic techniques are high speed data acquisition, off-line analysis capabilities that can connect to liquid chromatography (LC) separation, easy sample preparation with controllable sample consumption, and the ability to archive and recover samples [10][14][15][16][17]. Gadalla et al. compared flow cytometry with CyTOF and found that CyTOF was highly accurate, even in assessing low immune cell numbers [13]. From this investigation, they were able to develop a 40+ parameter panel for CyTOF which was used for broad scale immune cell profiling and biomarker discovery [13].
Proteomic techniques, however, are expensive, and access to a core with a mass spectrometer capable of running and analyzing proteomic data are not always available. Due to ion detection mechanisms, saturation effects, and signal to noise limitations, varied results are sometimes generated between instrumentation despite similar sample preparation protocols and instrument conditions [18]. Instrument variability can sometimes lead to proteins being masked in one system but be within the discoverable range in others.
2.3. Single Cell Sequencing
Immunology has greatly benefited from single cell sequencing which uses genomic techniques to evaluate the sequence of individual cells rather than the average of whole cell populations. Single cell sequencing evaluates the heterogeneity of cell populations to determine differences in genetics, maturity, or antigen presentation in addition to the host response, in turn giving more insight into how the immune system acts on an individual level within the host [19]. Because not all cells of a population have the same genetic sequence due to somatic mutations and methylation/epigenetic patterns introduced during DNA replication, single cell sequencing could lead to a boom of personalized medicine as it becomes more widely available and affordable [20].
Strengths and Limitations
One strength of single cell sequencing is the ability to perform multiomics analysis. Genomic, epigenetic, and transcriptomic data from the same cell set is more reliable than overlaying single data sets from multiple experiments due to fewer batch effects and decreased sampling bias [21]. One of the major challenges for single cell sequencing is that the initial isolation likely results in changes to cellular physiology. Cells are heavily influenced by their surrounding environment and cellular interactions. Single cells in suspension are no longer in their native environment, which could result in unintended cellular stress, altering the behavior, viability, and genetic profile of the cell [22]. Cell viability is required for downstream analysis, and new technologies are becoming available that allow for the isolation of cells in a gentle manner to ensure viability. These include cell printers and microfluidic platforms that harness picodroplet technology, which protect individual cells from shear stress.
Another caveat with single cell sequencing is the analysis of these data sets can be challenging and likely need a bioinformatician due to the nature of the multidimensional data generated [21]. In addition, single cell sequencing is more expensive than traditional bulk RNA sequencing. While single cell sequencing data on its own cannot provide spatial information, there are computational techniques that can be used to map out spatial data. Cellular clusters in single cell RNA-seq experiments, when coupled with immunohistochemistry staining, can provide important spatial information regarding the location of these cell types in the heart [21][23][24][25]. In addition, recent advancements have made it possible to determine spatial transcriptomics with as small as a 2 μm resolution by utilizing novel spatial barcoding technology on histological tissue sections for library preparation [26]. How well this new technology can capture some of the more minor populations such as T-cell subsets in the post-MI heart has not yet been studied.
References
- Strassheim, D.; Dempsey, E.C.; Gerasimovskaya, E.; Stenmark, K.; Karoor, V. Role of Inflammatory Cell Subtypes in Heart Failure. J. Immunol. Res. 2019, 2019, 2164017.
- Swirski, F.K.; Nahrendorf, M. Cardioimmunology: The immune system in cardiac homeostasis and disease. Nat. Rev. Immunol. 2018, 18, 733–744.
- Frangogiannis, N.G. The inflammatory response in myocardial injury, repair, and remodelling. Nat. Rev. Cardiol 2014, 11, 255–265.
- Yang, F.; Liu, Y.H.; Yang, X.P.; Xu, J.; Kapke, A.; Carretero, O.A. Myocardial infarction and cardiac remodelling in mice. Exp. Physiol. 2002, 87, 547–555.
- Zile, M.R.; Baicu, C.F.; Ikonomidis, J.S.; Stroud, R.E.; Nietert, P.J.; Bradshaw, A.D.; Slater, R.; Palmer, B.M.; Van Buren, P.; Meyer, M.; et al. Myocardial stiffness in patients with heart failure and a preserved ejection fraction: Contributions of collagen and titin. Circulation 2015, 131, 1247–1259.
- Jordan, A.R.; Racine, R.R.; Hennig, M.J.P.; Lokeshwar, V.B. The Role of CD44 in Disease Pathophysiology and Targeted Treatment. Front. Immunol. 2015, 6, 182.
- Cerovic, V.; Houston, S.A.; Westlund, J.; Utriainen, L.; Davison, E.S.; Scott, C.L.; Bain, C.C.; Joeris, T.; Agace, W.W.; Kroczek, R.A.; et al. Lymph-borne CD8alpha+ dendritic cells are uniquely able to cross-prime CD8+ T cells with antigen acquired from intestinal epithelial cells. Mucosal. Immunol. 2015, 8, 38–48.
- Jahan-Tigh, R.R.; Ryan, C.; Obermoser, G.; Schwarzenberger, K. Flow cytometry. J. Invest. Dermatol. 2012, 132, 1–6.
- Proserpio, V.; Lonnberg, T. Single-cell technologies are revolutionizing the approach to rare cells. Immunol. Cell Biol. 2016, 94, 225–229.
- Cramer, R. Maldi Ms. Methods Mol. Biol. 2009, 564, 85–103.
- Alexandrov, T. MALDI imaging mass spectrometry: Statistical data analysis and current computational challenges. BMC Bioinform. 2012, 13 (Suppl. S16), S11.
- Duncan, M.W.; Roder, H.; Hunsucker, S.W. Quantitative matrix-assisted laser desorption/ionization mass spectrometry. Brief Funct. Genomic. Proteomic. 2008, 7, 355–370.
- Gadalla, R.; Noamani, B.; MacLeod, B.L.; Dickson, R.J.; Guo, M.; Xu, W.; Lukhele, S.; Elsaesser, H.J.; Razak, A.R.A.; Hirano, N.; et al. Validation of CyTOF Against Flow Cytometry for Immunological Studies and Monitoring of Human Cancer Clinical Trials. Front. Oncol. 2019, 9, 415.
- Aichler, M.; Walch, A. MALDI Imaging mass spectrometry: Current frontiers and perspectives in pathology research and practice. Lab. Invest. 2015, 95, 422–431.
- Corr, J.J.; Kovarik, P.; Schneider, B.B.; Hendrikse, J.; Loboda, A.; Covey, T.R. Design considerations for high speed quantitative mass spectrometry with MALDI ionization. J. Am. Soc. Mass Spectrom. 2006, 17, 1129–1141.
- Mirgorodskaya, E.; Braeuer, C.; Fucini, P.; Lehrach, H.; Gobom, J. Nanoflow liquid chromatography coupled to matrix-assisted laser desorption/ionization mass spectrometry: Sample preparation, data analysis, and application to the analysis of complex peptide mixtures. Proteomics 2005, 5, 399–408.
- Brancia, F.L.; Bereszczak, J.Z.; Lapolla, A.; Fedele, D.; Baccarin, L.; Seraglia, R.; Traldi, P. Comprehensive analysis of glycated human serum albumin tryptic peptides by off-line liquid chromatography followed by MALDI analysis on a time-of-flight/curved field reflectron tandem mass spectrometer. J. Mass Spectrom. 2006, 41, 1179–1185.
- Michelle Byrd, H.C.; McEwen, C.N. The limitations of MALDI-TOF mass spectrometry in the analysis of wide polydisperse polymers. Anal. Chem. 2000, 72, 4568–4576.
- Tang, X.; Huang, Y.; Lei, J.; Luo, H.; Zhu, X. The single-cell sequencing: New developments and medical applications. Cell Biosci. 2019, 9, 53.
- Angermueller, C.; Clark, S.J.; Lee, H.J.; Macaulay, I.C.; Teng, M.J.; Hu, T.X.; Krueger, F.; Smallwood, S.; Ponting, C.P.; Voet, T.; et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat. Methods 2016, 13, 229–232.
- Kashima, Y.; Sakamoto, Y.; Kaneko, K.; Seki, M.; Suzuki, Y.; Suzuki, A. Single-cell sequencing techniques from individual to multiomics analyses. Exp. Mol. Med. 2020, 52, 1419–1427.
- Nguyen, Q.H.; Pervolarakis, N.; Nee, K.; Kessenbrock, K. Experimental Considerations for Single-Cell RNA Sequencing Approaches. Front. Cell Dev. Biol. 2018, 6, 108.
- Hu, Z.; Liu, W.; Hua, X.; Chen, X.; Chang, Y.; Hu, Y.; Xu, Z.; Song, J. Single-Cell Transcriptomic Atlas of Different Human Cardiac Arteries Identifies Cell Types Associated with Vascular Physiology. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 1408–1427.
- Molenaar, B.; Timmer, L.T.; Droog, M.; Perini, I.; Versteeg, D.; Kooijman, L.; Monshouwer-Kloots, J.; de Ruiter, H.; Gladka, M.M.; van Rooij, E. Single-cell transcriptomics following ischemic injury identifies a role for B2M in cardiac repair. Commun. Biol. 2021, 4, 146.
- Farbehi, N.; Patrick, R.; Dorison, A.; Xaymardan, M.; Janbandhu, V.; Wystub-Lis, K.; Ho, J.W.; Nordon, R.E.; Harvey, R.P. Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. Elife 2019, 8, e43882.
- Vickovic, S.; Eraslan, G.; Salmen, F.; Klughammer, J.; Stenbeck, L.; Schapiro, D.; Aijo, T.; Bonneau, R.; Bergenstrahle, L.; Navarro, J.F.; et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods 2019, 16, 987–990.
More
Information
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
642
Revisions:
2 times
(View History)
Update Date:
30 Dec 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





