Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Zeeshan Khan | + 2532 word(s) | 2532 | 2021-12-23 04:29:51 | | | |
| 2 | Catherine Yang | Meta information modification | 2532 | 2021-12-28 04:21:00 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Khan, Z. Antibiotic Susceptibility Testing. Encyclopedia. Available online: https://encyclopedia.pub/entry/17581 (accessed on 07 February 2026).
Khan Z. Antibiotic Susceptibility Testing. Encyclopedia. Available at: https://encyclopedia.pub/entry/17581. Accessed February 07, 2026.
Khan, Zeeshan. "Antibiotic Susceptibility Testing" Encyclopedia, https://encyclopedia.pub/entry/17581 (accessed February 07, 2026).
Khan, Z. (2021, December 28). Antibiotic Susceptibility Testing. In Encyclopedia. https://encyclopedia.pub/entry/17581
Khan, Zeeshan. "Antibiotic Susceptibility Testing." Encyclopedia. Web. 28 December, 2021.
Copy Citation
Antibiotic susceptibility testing (AST) specifies effective antibiotic dosage and formulates a profile of empirical therapy for the proper management of an individual patient’s health against deadly infections. Therefore, rapid diagnostic plays a pivotal role in the treatment of bacterial infection.
antibiotic susceptibility tests
resistance
1. Phenotypic AST Methods
1.1. Diffusion
The disk diffusion method is the gold standard for confirming the susceptibility of bacteria. Standardized disk diffusion was introduced by Bauer and Kirby’s experiments in 1956, after finalizing all aspects of optimization by changing physical conditions [1]. In this method, the isolated bacterial colony is selected, suspended into growth media, and standardized through a turbidity test. The standardized suspension is then inoculated onto the solidified agar plate, and the antibiotic-treated paper is tapped on the inoculated plate. The disc containing the antibiotic is allowed to diffuse through the solidified agar, resulting in the formation of an inhibition zone after the overnight incubation at 35 °C. Thereafter, the size of the inhibition zone formed around the paper disc is measured; the size of the inhibition zone corresponds to the concentration of antibiotic (Figure 1) [1][2]. Assessing and determining the susceptibility of bacteria generally takes 16–24 h. Several diffusion-based experiments have been performed prior to the standardized disk diffusion method. In the 1920s, Fleming was the pioneering contributor to AST. Fleming’s gutter method was the first method of antibiotic analysis where antibiotic was dispensed into a gutter made on solid agar that allowed the antibiotics to diffuse through it [3]. A modification to this design, called the “Oxford cup method,” was subsequently developed by Abraham et al. in 1941, where the gutter was replaced with a glass cup for diffusion [4]. Simultaneously, in the 1940s, Pope (1940), Foster and Woodruff (1943), and Vincet and Vincet (1944) used an antibiotic-impregnated paper disc for the diffusion of antibiotics [5][6]. These methods were hindered by inaccurate analysis due to evaporation, difficulty in handling, sterilization, and cumbersome operation [7]. A single antibiotic was focused on susceptibility testing (i.e., penicillin).
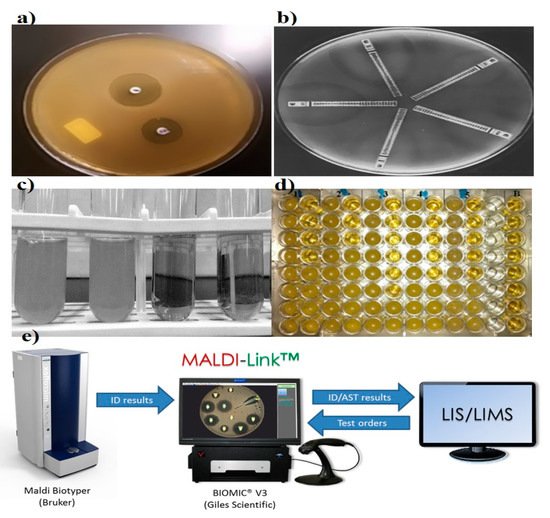
Figure 1. Representation of various conventional antibiotic susceptibility testing methods. (a) Disk diffusion, demonstrating of inhibition zones, adapted from Sageerabanoo [8]. (b) Etest gradient disk diffusion, adapted from Sader [9], under terms of the Creative Commons attribution license. (c,d) Broth macro and micro dilution, showing bacterial susceptibility based on optical density and (e) Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MADI-TOF MS), adapted from the MALDI Biotyper system (Bruker, Billerica, Massachusetts, United States), Laboratory Information System (LIS) and Laboratory Information Management System (LIMS).
Recently, the emergence of various instruments for analyzing the zone of inhibition has added to the reliability of the disk diffusion results by reducing variability due to operator handling and interpretation. The camera or scanner takes the picture, and the inbuilt image analysis software displays the zone of inhibition and compares the obtained results with the various guidelines present in the database. Accuzone (AccuMed International, Hillsboro, Oregon, USA), Biomic (Giles Scientific, Santa Barbara, California, USA), Mastascan Elite (Mast, Bootle, UK), and Sirscan (Becton Dickinson, Oxford, UK) are a few of the instruments capable of analyzing the zone of inhibition, but all differ in data input, analysis, ease of use, and presentation of results [10].
1.2. Dilution
Dilution was one of the earliest tools in microbiological practice, starting in the early 1870s, and it allows the growth and identification of bacterial populations in suspension [11]. Pasteur, Lister, Koch, and Ehrlich were listed as the pioneers in the field of bacteriology, and they worked on the concept of macrodilution [12]. William Roberts and John Tyndall further contributed to the macrodilution method and observed bacterial growth in a diluted medium [13]. The two basic types of dilution are microdilution and macrodilution, wherein broth and agar are the most commonly used mediums. In broth dilution, consecutive two-fold dilutions (1, 2, 4, 8, and 12 µL) of antibiotics are made and dispensed into micro-centrifuge tubes containing bacterial growth medium, followed by making up the final volume by adding the medium and incubating overnight at 35 °C. Finally, the growth examination is carried out for setting the breakpoint through the turbidity of culture media (Figure 1) [14][15]. In agar dilution, antibiotics are diluted into the agar medium, followed by plate formation and application of bacterial cells to the surface of the agar plate.
Microdilution is a miniaturized prototype of the macrodilution method where susceptibility testing is performed on disposable 96-well microtiter plates, where each well has a sample capacity of ~0.1 mL (Figure 1) [16]. To dispense the samples into microwells, mechanized dispensers are used to avoid the handling error. After overnight incubation, growth and MIC are assessed through specialized optical instruments. This method has been well standardized for most fastidious bacteria [2].
1.3. Etest
Epsilometer testing (Etest) is another significant development for the routine analysis of widespread antibiotic resistance in bacteria. In the late 1980s, Bolmström and Eriksson developed this test [17]. AB BIODISK manufactured the first Etest plastic strip to inspect multiple antibiotics on a single platform in 1991 (Figure 1) [17]. Etest plastic strips are coated with pre-defined antibiotic concentrations, and the corresponding interpretive MIC ranges are marked on the surface and back of the strip, respectively. For detection, multiple strips are placed on a pre-inoculated streaked agar plate, followed by an overnight incubation; elliptical inhibition zones appear around the strips, indicating the MIC at the intersection point between the inhibition zone and the strip edge [18]. The simplicity, accuracy, and reliability of the Etest makes it appropriate and convenient for Food and Drug Administration (FDA) approved commercialization [19]. The ability of convenient interpretations of MIC under diverse physical conditions made the Etest a preferential method over standardized disk diffusion and dilution techniques in clinical laboratories for AST [9][20].
Besides several advantages, there are some limitations that cannot be ignored, primarily related to the inaccurate and inconsistent behavior of the Etest for certain antibacterial agents, such as Penicillin, ciprofloxacin, ofloxacin, and rifampicin [21]. Some additional demerits that make the Etest complicated for routine test analyses are: pH-sensitive coated antibiotics, expensive batch performance, strip storage, and laboratory set-up for proper plate inoculation and incubation [22].
1.4. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS)
MALDI-TOF MS, introduced in 2000, is another sensitive method for bacterial identification. High sensitivity and accuracy are the key characteristics that make it a useful method for clinical relevance. Assorted studies reveal its significance in discriminating MRSA, MSSA, and other bacterial strains where susceptible and resistant bacteria have been evaluated through spectral peak analysis. Even the subtle difference in expression profiles have been noticed in isogenic strains of S. aureus [23][24]. The efficiency of MALDI-TOF MS has been further investigated on vancomycin-resistant Enterococci, where sensitivity higher than 90% has been recorded. Furthermore, analysis of multiple targets with different resistant strains of Pseudomonas spp. against ciprofloxacin, tobramycin, and meropenem have been identified efficiently [25]. The newly developed MALDI Biotyper antibiotic susceptibility test rapid assay (MBT-ASTRA) is a more-straightforward and cost-effective modulation of MALDI-TOF MS used for both AST and MIC determination [26]. Despite all the advantage of MALDI-TOF MS, the expensive nature of the instrument and its maintenance are prime disadvantages for mass application.
1.5. Automated Systems
Since the dawn of automated technologies in the 1980s, antibiotic susceptibility tests have been perpetually improvised and, hence, have superseded conventional phenotypic methods [2]. Automation, simplicity, and compactness are the major reasons for their widespread acceptance in diagnostics. Computer integration has allowed online analysis and data sharing, which is a giant leap for results validation, especially in remote areas [27]. Among the developed automated systems, MicroScan WalkAway (Beckman Coulter, Inc. Atlanta, Georgia, USA) (1980), Micronaut (Merlin, berlin, Germany) (1990), the avantage test (Abbott Laboratories, Irving, Texas, USA) (1980), Vitek 2 (bioMe’rieux, Marcy-l’Étoile, France) (2000), Phoenix (BD Diagnostics, Franklin Lakes, New jersey, USA) (2001), and Sensititre ARIS 2X (Trek Diagnostic Systems, Oakwood Village, Ohio, USA) (2004) are the major FDA approved systems for AST. Vitek and Pheonix detect growing bacteria on the basis of turbidity, whereas comparable automated systems like MicroScan WalkAway (Beckman Coulter, Inc. Atlanta, Georgia, USA) and Sensititre ARIS 2X (are based on fluorescence emission of the growing bacteria. The resistance in gram-negative, gram-positive, and Streptococcus strains of bacteria can easily be estimated through Phoenix, and Vitek 2, but, MicroScan WalkAway and Sensititre ARIS 2XESBL (Trek Diagnostic Systems, Oakwood Village, Ohio, USA) are capable of detecting the extended-spectrum beta-lactamase (ESBL)-producing strains in the species mentioned above [28]. Micronaut and Avantage are capable of accurate direct susceptibility testing for gram-positive and gram-negative bacteria, respectively [29][30].
Detection incompatibility for many bacteria/antibiotics using Sensititre ARIS 2 and Vitek 1 have led to the development of the updated Sensititre ARIS 2X and Vitek 2, which have better performances and are applicable to a broader range of bacteria/antibiotics. Presently, all the automated systems are incorporated with advanced expert system software for enhanced performance and online data processing [10]. Every automated system has a specific panel capacity and an average time for performing detection, which varies from 40 to 100 wells, and can vary in time such as 20, 12 and 9 h, respectively. Certain models such as Phoenix AP, and Vitek 2Xl, are dedicated towards automated inoculation, enhanced card capacity and compactness.
The aforementioned systems lack reproducibility, sensitivity, and reliability compared with the existing traditional methods. Moreover, an inability to test a wide range of clinically relevant bacteria (e.g., S. pneumonia), antimicrobial agents (e.g., vancomycin), and heteroresistant isolates, as well as a limited panel capacity and the high cost of instruments and consumables, are all significant issues that restrict these systems from frequent analysis [31].
2. Genotypic AST Methods
Molecular or genotypic AST are the effective direct methods that eliminate tedious bacterial cultures, long incubation, chances of contamination, and the spreading of deadly infections [32]. PCR, DNA microarray and DNA chips, and loop-mediated isothermal amplification (LAMP) are some of the genotypic techniques for the detection of antibiotic resistance. Mutational assessment of methicillin resistance in Staphylococcus spp., vancomycin resistance in Enterococcus spp., and multi-antibiotic (isoniazid, rifampin, streptomycin, pyrazinamide, and the fluoroquinolones) resistance in Mycobacterium spp. have been successfully estimated through various genotypic techniques.
PCR is one of the most efficient and rapid molecular tools for quantification and profiling of bacterially infectious genes. The first report on PCR diagnostic application was published by Saiki et al. [33]. The general methodology of PCR includes cycles of denaturation, annealing of the primers, and elongation of the primers by a thermostable DNA polymerase in a compatible buffer containing nucleotides, ions, and so on. Each cycle of amplification doubles the target DNA molecule. The amplified target can be confirmed for the presence of resistance genes through electrophoresis, southern blotting, restriction fragment-length polymorphism, single-strand conformation polymorphism (SSCP), DNA fingerprinting, molecular beacons, and other DNA sequencing analysis methods (Figure 2) [33][34]. Another tool developed on the basis of PCR is LAMP, which has also been used for the evaluation of AST. In LAMP, the gene of interest is amplified at a constant temperature of 60–65 °C using a Bst DNA polymerase instead of Taq polymerase because of strong strand displacement activity (required in isothermal techniques) [35].
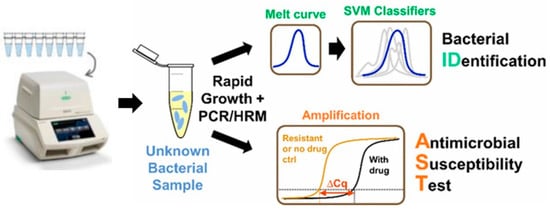
Figure 2. Digital PCR-High Resolution Melt analysis (HRM)-based bacterial identification from mixed bacterial samples, reproduced with permission from [36], published by American Chemical Society, 2017. (SVM: Support-vector machine)
DNA microarrays and DNA chips are the other promising technologies utilized for screening susceptibility [37]. DNA arrays employ cDNA fragment probes on nylon membrane, where each DNA chip has a glass or silicon platform for probe binding. The specific hybridization of the labeled probe with the target and its recognition help to determine the resistance. Determination of isoniazid resistance in M. tuberculosis has been carried out successfully through DNA microarrays and chips [38][39]. Colorimetric detection and multiplexing are the attractive features of these techniques.
3. Emerging Methods for AST
Microfluidics-based diagnostics are one of the most promising emerging tools for AST. Microfluidics is an evolving field characterized by the manipulation of fluids in micro-volume, thereby offering portability, cost-effectiveness, multiplexing, reproducibility, and a controllable environment in an in vitro system [1]. The concept was first introduced in the semiconductor and micro-electromechanical systems (MEMS) industries, then further extended to the field of biomedical research [40]. Integrated microfluidic devices, generally referred to as micro total analysis systems (µTAS), are proficient in performing molecular diagnostics [41].
The quantity of samples has always been the biggest challenge for biological studies in general, and pathological analysis in particular. Since microfluidics is capable of dealing with the minimal quantity of samples, it has therefore emerged as a promising tool for pathologists. Currently, microfluidics platforms are capable of single-cell analysis, and can even analyze the single-cell interrogation of signaling networks in cultured cell lines. In the past decades, numerous conventional and automated strategies have been successfully embraced to diagnose the ever-increasing antibiotic resistance in bacteria [42]. Nevertheless, the most-practiced techniques, such as disk diffusion and microdilution, have a high chance of cross-contamination, laborious protocols, lengthy processing times, improper power supplies, and cumbersome set-ups that render their application in resource-limited regions, and unfortunately, these regions have a higher rate of resistance too [43][44]. The rise of microfluidics as a diagnostic tool has shown promise in addressing the abovementioned shortcomings [45][46].
Progression in optical imaging has developed various image sensors with high sensitivity and resolution that are capable of biological analysis. These imaging systems are frequently used for morphological and growth studies of bacteria. Generally, owing to real-time analysis and minimum culture dependency, microfluidic devices coupled with an optical sensor can perform AST and detect the MIC in few hours [47]. Recent reports based on single bacterial cell analysis have claimed that optical sensor-based nanofluidic (30 nl) can finish AST within 30 min [48]. This direct imaging of single bacterium requires simple sample preparation steps, but eliminates the tedious steps of continuous sample injection, loading of cells, and counting-based cell identification [48].
Fluorescence proteins and dyes are commonly used for tagging resistant biomarkers. These proteins/dyes can be of biological or chemical origin. One of the pioneer proteins used for imaging is green fluorescent protein (GFP). GFP, obtained from jellyfish Aequoria victoria, is essential for the noninvasive real-time monitoring of antimicrobial susceptibility [49]. Adding to this, the same group was able to evaluate multiple antibiotic sensitivities in real-time in polymicrobial culture bacterial strains (namely, E. coli, P. aeruginosa, and K. pneumoniae) simultaneously. Green and red fluorescent protein tagging have been utilized for real-time growth quantification in the presence of multiple antibiotics on a multiplexed microfluidic platform (Figure 3) [50]. All these methods are sensitive and reliable, but creating these recombinant bacteria requires molecular handling, which is a challenge for routine clinical observations. Fluorescent dyes are another means for optical fluorescence tagging. These chemicals, with all the benefits of the fluorescence protein, can avoid stearic hindrance due to their small size, and elimination of cloning or transformation. SYTOX green and resazurin (alamarBlue; inactive precursor of resazurin) are two common fluorescence indicator dyes used for viability testing in AST [51][52]. Similarly, resazurin can be utilized in colorimetric detection. When the culture media is supplemented with antibiotic and resazurin, a uniform intense blue color is obtained in the beginning. In the presence of resistance, the resazurin is reduced to resafurin, and the intense blue color changes to pink and leuco; in the absence of resistance in bacteria, the blue color sustains. This method is also translated to a microfluidic chip for MIC estimation for four different antibiotics against 20 clinical strains of Escherichia and Shigella [52].
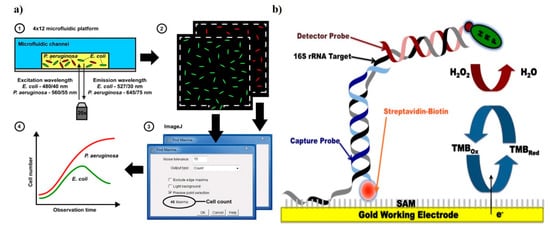
Figure 3. Demonstration of sensors for antibacterial susceptibility testing involving (a) an optical microfluidics biosensor, showing optical detection of microbial cultures, reproduced with permission from [50], published by RCS Advances, 2015; and (b) an electrochemical biosensor, detection was based on hybridization of the target bacterial 16S rRNA with a detector probe, adapted with permission from Liu [53] (under terms of the Creative Commons attribution license).
References
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496.
- Reller, L.B.; Weinstein, M.; Jorgensen, J.H.; Ferraro, M.J. Antimicrobial Susceptibility Testing: A Review of General Principles and Contemporary Practices. Clin. Infect. Dis. 2009, 49, 1749–1755.
- Fleming, A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Br. J. Exp. Pathol. 1929, 10, 226.
- Abraham, E.P.; Chain, E.; Fletcher, C.M.; Gardner, A.D.; Heatley, N.G.; Jennings, M.A.; Florey, H.W. Further observations on penicillin. Lancet 1941, 238, 177–189.
- Foster, J.W.; Woodruff, H.B. Microbiological aspects of penicillin: I. Methods of assay. J. Bacteriol. 1943, 46, 187.
- Vincent, J.G.; Vincent, H.W.; Morton, J. Filter Paper Disc Modification of the Oxford Cup Penicillin Determination. Proc. Soc. Exp. Biol. Med. 1944, 55, 162–164.
- Gavin, J.J. Analytical Microbiology: II. The Diffusion Methods. Appl. Microbiol. 1957, 5, 25.
- Sageerabanoo, S.; Malini, A.; Mangaiyarkarasi, T.; Hemalatha, G. Phenotypic detection of extended spectrum β-lactamase and Amp-C β-lactamase producing clinical isolates in a Tertiary Care Hospital: A preliminary study. J. Nat. Sci. Biol. Med. 2015, 6, 383–387.
- Sader, H.S.; Pignatari, A.C. E test: A novel technique for antimicrobial susceptibility testing. Sao Paulo Med. J. Rev. Paul. Med. 1994, 112, 635–638.
- Felmingham, D.; Brown, D.F. Instrumentation in antimicrobial susceptibility testing. J. Antimicrob. Chemother. 2001, 48, 81–85.
- Guardino, R.F. Early History of Microbiology and Microbiological Methods; Parenteral Drug Association: Wilmington, DE, USA, 2005.
- Rittenberg, S.C. Three Centuries of Microbiology. Hubert, A. Lechevalier and Morris Solotorovsky. McGraw-Hill, New York, 1965. viii + 536 pp. Paper, $4.95. Science 1965, 149, 530–531.
- Poupard, J.A.; Rittenhouse, S.F.; Walsh, L.R. The evolution of antimicrobial susceptibility testing methods. In Antimicrobial Susceptibility Testing; Springer, Plenum Publishing: New York, NY, USA, 1994; pp. 3–14.
- Ericsson, H.M.; Sherris, J.C. Antibiotic sensitivity testing. Report of an international collaborative study. Acta Pathol. Microbiol. Scand. 1971, 90.
- Jorgensen, J.H.; Turnidge, J.D.; Washington, J.A.; Murray, P.R.; Pfaller, M.A.; Tenover, F.C.; Baron, E.J.; Yolken, R.H. Antibacterial susceptibility tests: Dilution and disk diffusion Methods. In Manual of Clinical Microbiology; Geo. F. Brooks Publisher: Washington, DC, USA, 1999.
- Tang, Y.-W.; Stratton, C.W. Advanced Techniques in Diagnostic Microbiology; Springer: Nashville, TN, USA, 2012.
- Picard, J. Applied Veterinary Bacteriology and Mycology: Bacteriological Techniques; University of Pretoria, Afrivip: Pretoria, South Africa, 1990.
- Sanchez, M.L.; Jones, R.N. E test, an antimicrobial susceptibility testing method with broad clinical and epidemiologic application. Antimicrob. Newsl. 1992, 8, 1–7.
- Joyce, L.F.; Downes, J.; Stockman, K.; Andrew, J.H. Comparison of five methods, including the PDM Epsilometer test (E test), for antimicrobial susceptibility testing of Pseudomonas aeruginosa. J. Clin. Microbiol. 1992, 30, 2709–2713.
- Baker, C.N.; Stocker, S.A.; Culver, D.H.; Thornsberry, C. Comparison of the E Test to agar dilution, broth microdilution, and agar diffusion susceptibility testing techniques by using a special challenge set of bacteria. J. Clin. Microbiol. 1991, 29, 533–538.
- Frosch, M.; Maiden, M.C.J. Handbook of Meningococcal Disease: Infection Biology, Vaccination, Clinical Management; John Wiley & Sons: Hoboken, NJ, USA, 2006.
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79.
- Bernardo, K.; Pakulat, N.; Macht, M.; Krut, O.; Seifert, H.; Fleer, S.; Hünger, F.; Krönke, M. Identification and discrimination of Staphylococcus aureus strains using matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Proteomics 2002, 2, 747–753.
- Hrabák, J.; Chudáčková, E.; Walková, R. Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry for detection of antibiotic resistance mechanisms: From research to routine diagnosis. Clin. Microbiol. Rev. 2013, 26, 103–114.
- Jung, J.; Eberl, T.; Sparbier, K.; Lange, C.; Kostrzewa, M.; Schubert, S.; Wieser, A. Rapid detection of antibiotic resistance based on mass spectrometry and stable isotopes. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 949–955.
- Zimmermann, S.; Burckhardt, I. Development and Application of MALDI-TOF for Detection of Resistance Mechanisms. In MALDI-TOF and Tandem MS for Clinical Microbiology; John Wiley & Sons: Hoboken, NJ, USA, 2017; pp. 231–248.
- Richter, S.S.; Ferraro, M.J. Susceptibility testing instrumentation and computerized expert systems for data analysis and interpretation. In Manual of Clinical Microbiology, 10th ed.; American Society of Microbiology: Sterling, VA, USA, 2011; pp. 1144–1154.
- Sellenriek, P.; Holmes, J.; Ferrett, R.; Drury, R.; Storch, G.A. Comparison of MicroScan Walk-Away®, Phoenix™ and VITEK-TWO® Microbiology systems used in the identification and susceptibility testing of bacteria. In Proceedings of the Abstr 105th General Meeting of the American Society for Microbiology, Atlanta, GA, USA, 5–9 June 2005.
- Wright, D.N.; Matsen, J.M.; DiPersio, J.R.; Kirk, M.; Saxon, B.; Ficorilli, S.M.; Spencer, H.J. Evaluation of four newer antimicrobial agents in the Avantage susceptibility test system. J. Clin. Microbiol. 1989, 27, 2381–2383.
- Wellinghausen, N.; Pietzcker, T.; Poppert, S.; Belak, S.; Fieser, N.; Bartel, M.; Essig, A. Evaluation of the Merlin MICRONAUT System for Rapid Direct Susceptibility Testing of Gram-Positive Cocci and Gram-Negative Bacilli from Positive Blood Cultures. J. Clin. Microbiol. 2007, 45, 789–795.
- Karlowsky, J.A.; Richter, S.S. Antimicrobial susceptibility testing systems. In Manual of Clinical Microbiology, 11th ed.; American Society of Microbiology: Sterling, VA, USA, 2015; pp. 1274–1285.
- Cockerill, F.R. Genetic methods for assessing antimicrobial resistance. Antimicrob. Agents Chemother. 1999, 43, 199–212.
- Fluit, A.C.; Visser, M.R.; Schmitz, F.-J. Molecular detection of antimicrobial resistance. Clin. Microbiol. Rev. 2001, 14, 836–871.
- Miller, M.B.; Tang, Y.-W. Basic Concepts of Microarrays and Potential Applications in Clinical Microbiology. Clin. Microbiol. Rev. 2009, 22, 611–633.
- Li, Y.; Fan, P.; Zhou, S.; Zhang, L. Loop-mediated isothermal amplification (LAMP): A novel rapid detection platform for pathogens. Microb. Pathog. 2017, 107, 54–61.
- Athamanolap, P.; Hsieh, K.; Chen, L.; Yang, S.; Wang, T.H. Integrated Bacterial Identification and Antimicrobial Susceptibility Testing Using PCR and High-Resolution Melt. Anal. Chem. 2017, 89, 11529–11536.
- Frye, J.G.; Lindsey, R.L.; Rondeau, G.; Porwollik, S.; Long, F.; McClelland, M.; Jackson, C.R.; Englen, M.D.; Meinersmann, R.J.; Berrang, M.E.; et al. Development of a DNA microarray to detect antimicrobial resistance genes identified in the National Center for Biotechnology Information database. Microb. Drug Resist. 2010, 16, 9–19.
- Gryadunov, D.; Mikhailovich, V.; Lapa, S.; Roudinskii, N.; Donnikov, M.; Pan’kov, S.; Markova, O.; Kuz’min, A.; Chernousova, L.; Skotnikova, O.; et al. Evaluation of hybridisation on oligonucleotide microarrays for analysis of drug-resistant Mycobacterium tuberculosis. Clin. Microbiol. Infect. 2005, 11, 531–539.
- Huang, W.L.; Hsu, Z.J.; Chang, T.C.; Jou, R. Rapid and accurate detection of rifampin and isoniazid-resistant Mycobacterium tuberculosis using an oligonucleotide array. Clin. Microbiol. Infect. 2014, 20, O542–O549.
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189.
- Park, S.; Zhang, Y.; Lin, S.; Wang, T.H.; Yang, S. Advances in microfluidic PCR for point-of-care infectious disease diagnostics. Biotechnol. Adv. 2011, 29, 830–839.
- Ibrahim, W.A.; Marouf, S.A.; Erfan, A.M.; Nasef, S.A.; Jakee, J.K.E. The occurrence of disinfectant and antibiotic-resistant genes in Escherichia coli isolated from chickens in Egypt. Vet. World 2019, 12, 141–145.
- Martínez, J.L. Antibiotics and Antibiotic Resistance Genes in Natural Environments. Science 2008, 321, 365–367.
- Ayukekbong, J.A.; Ntemgwa, M.; Atabe, A.N. The threat of antimicrobial resistance in developing countries: Causes and control strategies. Antimicrob. Resist. Infect. Control 2017, 6, 47.
- Chin, C.D.; Laksanasopin, T.; Cheung, Y.K.; Steinmiller, D.; Linder, V.; Parsa, H.; Wang, J.; Moore, H.; Rouse, R.; Umviligihozo, G.; et al. Microfluidics-based diagnostics of infectious diseases in the developing world. Nat. Med. 2011, 17, 1015–1019.
- Martinez, A.W.; Phillips, S.T.; Whitesides, G.M.; Carrilho, E. Diagnostics for the Developing World: Microfluidic Paper-Based Analytical Devices; ACS Publications: Sterling, VA, USA, 2009.
- Chen, C.H.; Lu, Y.; Sin, M.L.Y.; Mach, K.E.; Zhang, D.D.; Gau, V.; Liao, J.C.; Wong, P.K. Antimicrobial susceptibility testing using high surface-to-volume ratio microchannels. Anal. Chem. 2010, 82, 1012–1019.
- Baltekin, Ö.; Boucharin, A.; Andersson, D.I.; Elf, J. Fast Antibiotic Susceptibility Testing (FASTest) based on single cell growth rate measurements. bioRxiv 2016.
- Webb, J.S.; Barratt, S.R.; Sabev, H.; Nixon, M.; Eastwood, I.M.; Greenhalgh, M.; Handley, P.S.; Robson, G.D. Green fluorescent protein as a novel indicator of antimicrobial susceptibility in Aureobasidium pullulans. Appl. Environ. Microbiol. 2001, 67, 5614–5620.
- Mohan, R.; Sanpitakseree, C.; Desai, A.V.; Sevgen, S.E.; Schroeder, C.M.; Kenis, P.J.A. A microfluidic approach to study the effect of bacterial interactions on antimicrobial susceptibility in polymicrobial cultures. RSC Adv. 2015, 5, 35211–35223.
- Kalashnikov, M.; Lee, J.C.; Campbell, J.; Sharon, A.; Sauer-Budge, A.F. A microfluidic platform for rapid, stress-induced antibiotic susceptibility testing of Staphylococcus aureus. Lab Chip 2012, 12, 4523–4532.
- Elavarasan, T.; Chhina, S.K.; Ash, M.P.; Sankaran, K. Resazurin reduction based colorimetric antibiogram in microfluidic plastic chip. Sens. Actuators B Chem. 2013, 176, 174–180.
- Liu, T.; Lu, Y.; Gau, V.; Liao, J.C.; Wong, P.K. Rapid antimicrobial susceptibility testing with electrokinetics enhanced biosensors for diagnosis of acute bacterial infections. Ann. Biomed. Eng. 2014, 42, 2314–2321.
More
Information
Subjects:
Infectious Diseases
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.9K
Revisions:
2 times
(View History)
Update Date:
03 Jan 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




