Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Robert Neil Kelsh | + 3721 word(s) | 3721 | 2021-12-20 07:25:41 | | | |
| 2 | Jason Zhu | Meta information modification | 3721 | 2021-12-24 10:05:40 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Kelsh, R. Neural Crest Cells. Encyclopedia. Available online: https://encyclopedia.pub/entry/17547 (accessed on 07 February 2026).
Kelsh R. Neural Crest Cells. Encyclopedia. Available at: https://encyclopedia.pub/entry/17547. Accessed February 07, 2026.
Kelsh, Robert. "Neural Crest Cells" Encyclopedia, https://encyclopedia.pub/entry/17547 (accessed February 07, 2026).
Kelsh, R. (2021, December 24). Neural Crest Cells. In Encyclopedia. https://encyclopedia.pub/entry/17547
Kelsh, Robert. "Neural Crest Cells." Encyclopedia. Web. 24 December, 2021.
Copy Citation
The neural crest shows an astonishing multipotency, generating multiple neural derivatives, but also pigment cells, skeletogenic and other cell types. Here w consider how these multipotent cells may give rise to all those diverse cell-types.
neural crest cells
peripheral nervous system
sensory neuron
1. Introduction
Neural crest cells (NCCs) contribute substantially to the formation of the peripheral nervous system, providing sensory neurons within the dorsal root ganglia, enteric neurons throughout the gut, and sympathetic neurons, as well as the accompanying glial fates, including Schwann cells and satellite glia [1]. However, NCCs also generate a remarkable diversity of other cell types, including cartilage and bone, smooth muscle, adrenal medullary cells, and numerous pigment cells [1]. Early studies showed that many (and presumably all?) early neural crest cells are multipotent, and they are widely considered to at least start off that way [2][3][4][5][6]. A long-standing debate (going back to the 1980s) concerns the mechanism whereby individual fates are chosen. Proponents of the direct fate restriction (DFR) mechanism argue that all fates are directly assigned from fully multipotent cells, whereas others have proposed the progressive fate restriction (PFR) model in which individual fates are selected through a series of intermediate progenitors in which a subset of fate options are retained, but all others are lost (Figure 1a,b) [7][8][9][10][11][12][13][14][15]. These two views have sat uncomfortably together, and have never been well resolved, although PFR has become strongly favored, especially in the context of the peripheral nervous system, where apparently bipotent progenitors for neurons and glial components of sensory and sympathetic ganglia have been characterized.
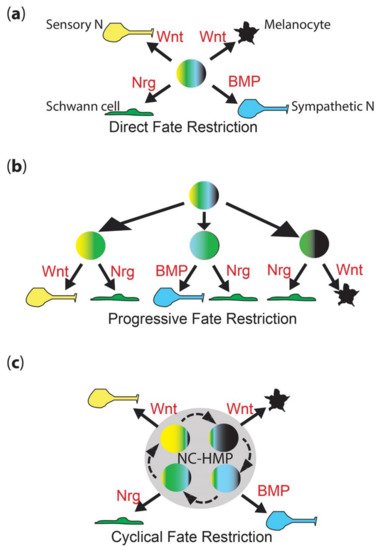
Figure 1. Neural crest fate restriction models. (a) Direct fate restriction (DFR). In the DFR model, highly multipotent NCC (rainbow colored) generates each fate directly (solid arrow), under direct influence of environmental signals (shown in red). Work in mammals suggests that Wnt signaling is important for both melanocyte and sensory neuron fate specification, with differences in signal timing being key [16]. (b) Progressive fate restriction (PFR). In the PFR model, multipotent NCC rapidly generates a series of intermediate progenitors whose options are restricted. In this schematic, for simplicity, they are shown only as the best-characterized bipotent intermediates (two color), but these models also posit higher potency intermediates as well. The partially restricted (here, bipotent) intermediates then generate individual fates under the influence of environmental signals (red). (c) New cyclical fate restriction (CFR) model. In the novel CFR model, NCCs enter a highly multipotent progenitor state (NC-HMP, grey circle), characterized by a highly dynamic transcriptome, which cycles (dashed arrows) through a series of sub-states; note that this state is highly multipotent (rainbow colors), but each sub-state is biased towards an individual fate (broader band of one color); under the influence of environmental signals (red), cells in each sub-state are driven to adopt an individual fate (solid arrow). Note that these simplified schematics focus on only four key fates—melanocyte (black), Schwann cell (green), sensory neuron (yellow) and sympathetic neuron (cyan)—and on key features of each model.
2. Classical Views of Neural Crest Development
2.1. Direct Fate Restriction
Pioneering experiments by Marianne Bronner using iontophoretic dye labelling of neural plate and delaminated NCCs performed on chick embryos observed a mixture of clones contributing to two or more cell types, including peripheral neural, pigment and other fates [13][14][15]. These were interpreted as indicating that neural crest cells might be initially highly multipotent, and then choose fate in accordance with environmental cues in the target destination where differentiation occurs. Thus, cells were considered to migrate in a fully multipotent state. The identification and characterization of neural crest stem cells (NCSCs) by David Anderson’s lab reinforced this view, showing in culture that different environmental cues (growth factors, including BMPs, Neuregulin) drove differentiation of sensory and sympathetic neurons, Schwann cell and smooth muscle fates [17]. Importantly, careful clonal studies demonstrated that the process was instructive, not selective, i.e., the growth factors direct the differentiation of the NCSCs, rather than simply selecting against those that had made the wrong decision through some other mechanism [18][19][20][21][22][23]. Detailed characterization of the process in neuronal development showed the growth factor-dependent transcriptional activation of fate-specific transcription factors was an important initial step in differentiation: thus, for sympathetic neuron development, BMP signaling induces and maintains expression of MASH1 and PHOX2B, whereas for sensory neuron development, Wnt signaling drives Neurogenin1/2 expression [19][20][21][22][23]. Similarly, for Schwann cell fate, both GGF and Notch signaling were crucial, although the key transcription factors affected have remained less clear, with SOX10 important, but with a complex role being involved in neuronal fates too [18][23][24]. Thus, the direct fate restriction (DFR) model posits that fully multipotent cells undergo environmental signal-driven specification directly to individual fates (Figure 1a). When originally proposed, the model was closely linked to the idea that fate specification occurs after migration, consistent with the in vivo observation of high BMP expression in the dorsal aorta adjacent to the site of sympathetic ganglial development [19][25].
2.2. Progressive Fate Restriction
Nicole Le Douarin, whose group had observed extensive heterogeneity in clonal composition of primary neural crest cells cultured under conditions conducive to differentiation of diverse cell types, and Jim Weston, noting the heterogeneity of marker expression in premigratory and early migrating neural crest, both suggested an alternative model, progressive fate restriction (PFR; Figure 1b) [7][8][9][10][11][12]. They suggested that initially highly multipotent neural crest cells underwent a series of partial restrictions in their potency, resulting eventually in adoption of a single fate. Again, this was integrated with NCC migration, with the suggestion that the process began prior to or during migration, so that cells arriving at their terminal destination might already be restricted to a subset of fates—and indeed that their migration pathways might be controlled by these restriction decisions. Thus, for example, cells restricted to sympathetic neuron and glial fates (bipotent sympathetic neuroglioblasts) might be guided to the vicinity of the dorsal aorta, forming the nascent sympathetic ganglia; cells that became partially restricted to sensory neuron and glial fate (bipotent sensory neuroglioblasts) would, instead, be directed to the vicinity of the spinal cord. This would be consistent with the striking regularity of NCC migration (but see also more recent evidence arguing that migration in the trunk is rather more random; [26]) and, for melanoblasts on the dorsolateral migration pathway, has been dissected molecularly in chick [27][28][29][30][31][32][33]. The final selection of neuronal versus glial fates would, in each case, result from a local communication process involving both ErbB/neuregulin signaling and Notch-delta lateral inhibition processes [18][24].
3. Conflicting Observations within Recent Studies
Although widely debated in the late 20th century, the discussion became less prominent subsequently, perhaps because the PFR model became favored. Nevertheless, the issue remained unresolved, and at least one key developmental biology textbook makes reference to both sets of studies, and both ideas, without attempting to resolve them [34]. However, the issue is important, not least because if NCCs work through a DFR mechanism, that is rather distinct from the way researchers tend to think about development in general. A careful modern clonal fate-mapping study in mice concluded that even migratory NCCs retained multipotency, but in the context of the PFR versus DFR debate that is the focus, it defined multipotency as ‘fated to form at least two cell-types’, a definition that does not distinguish between PFR and DFR [35]. A recent tour de force study of mouse and chick NC development using scRNA-seq generated a molecular representation of the PFR model, with neural development resulting from a series of ‘sequential binary decisions’ with initial coactivation of both programs followed by commitment to one and repression of the other; they describe early segregation of sensory neural progenitors (from which are generated sensory neurons and glia), with other cells then choosing between mesenchymal and autonomic neural fates, and the latter finally choosing between neuronal and glial commitment [36]. Similar conclusions regarding early segregation of neuronally, neuraly and mesenchymally biased progenitors were reached in the context of vagal NC development in chick [37].
3.1. Early Fate Specification and Ongoing Multipotency
Examination of key fate-specific markers has expanded the observations of heterogeneity that Weston noted, indicating that many NCCs show evidence of fate specification at premigratory and migratory stages (e.g., [38][39][40][41][42][43][44][45]). Conversely, the identification of NCSCs, and their isolation from multiple locations throughout the body, indicated that at least some individual NCCs retained multipotency during migration and in their post-migratory locations (reviewed in [46]). Some of these cells, most notably those (Melanocyte Stem Cells) giving rise to melanocytes in the adult skin, were initially assumed to be unipotent, but further investigation in mammals showed they had latent multipotency [47][48]. This begs the question of whether these cells are somehow different from the beginning, or is it their final location (the niche) that enables them to retain a cryptic multipotency? Study of pigment cell development in zebrafish embryos has illuminated both these aspects of NCC development.
3.2. Zebrafish Pigment Cell Development
Study of NCCs in zebrafish has revealed a strong conservation of derivative cell types and the key genetic factors in their fate specification. In the context of the peripheral nervous system, roles for Neurogenin1, ErbB, Phox2bb, and Sox10 in fate specification of sensory neuron, sympathetic and enteric neuron and glial fates have been shown, paralleling their roles in mammals [49][50][51][52][53][54][55][56]. In the context of the PFR model, bipotent neuroglioblasts have been deduced, with studies of neurog1 mutants and morphants (embryos generated by injection of morpholinos, and usually showing mosaic knockdown phenotypes) showing that sensory neuroglioblasts adopt glial fates when Neurog1 function inhibited [49].
Particular attention has been focused on the highly visible pigment cell derivatives, partly because zebrafish have melanocytes (often referred to as melanophores) just like mammals, and partly because an abundance of mutants has generated a significant genetic resource for study (e.g., [42][45][57][58][59][60][61][62][63]). Important early work showed that Mitfa, one (of two) zebrafish orthologue of mammalian Mitf, played a pivotal role in melanocyte fate specification from NCCs, with all NC-derived melanocytes being absent in strong loss-of-function mutants, and with expression in the NC being sufficient to rescue mitfa mutants [45][64]. Likewise, the role for Sox10 in driving mitfa expression has been well demonstrated [65], showing that the role for Sox10 in fate specification of melanocytes is conserved.
In the context of the fate restriction debate, the real interest in zebrafish stems from the fact that, in fish, there are usually multiple types of pigment cells [66]. In the zebrafish embryo, these consist of melanocytes, but also silver iridophores and yellow xanthophores. Single cell iontophoretic labelling studies have clearly shown that all these pigment cells are NCC derived [53][67][68], whilst genetic studies have identified at least some of the key transcription factors and growth factors and their receptors that are important in their fate specification [39][40][41][42][44][69][70][71][72][73][74]. An influential theory from Joe Bagnara [75] proposed that all pigment cells share a common developmental origin, exclusive to these fates—a classical partially fate restricted progenitor, which researchers will name a chromatoblast. This idea remained untested, but the emergence of the zebrafish model with its multiple cell fates and genetic tractability, finally opened up the possibility. In addition, a key observation from studies of the conserved role of Mitfa is that the absence of melanocytes is accompanied by a substantial increase in the number of iridophores [45]. This observation, plus similar observations in the context of other pigment cell types in another fish, medaka, were clearly consistent with the idea of bipotent pigment cell progenitors (i.e., here, a melanoiridoblast that forms both melanocytes and iridophores) [76][77][78]. The fit to a PFR model, with the chromatoblast being a more multipotent progenitor of the bipotent melanoiridoblast, was hard to ignore, and led to widespread assumption, by us and by others in the field, that a PFR model would explain zebrafish pigment cell development (Figure 2). Such a view formed the interpretative framework for a number of recent single cell RNA-seq studies, which have identified pigment cell progenitors (putative chromatoblasts) [60][79][80]; only in one case have they been assigned to a melanoiridoblast type, but these cells are from very early larval stages and here there is low-level expression of pax7b (a xanthophore lineage marker) too, suggesting they might have higher potency [80].
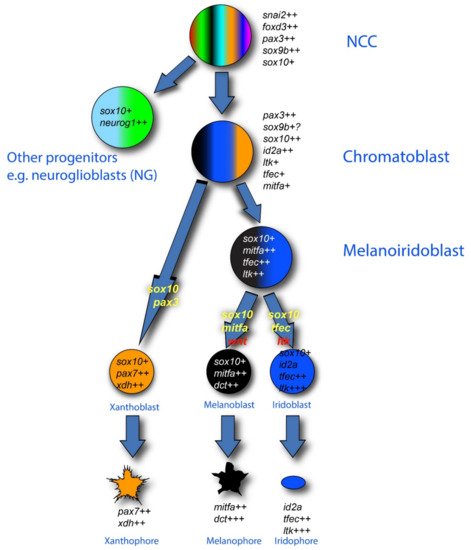
Figure 2. Pigment cell development in zebrafish: conventional PFR model. The three zebrafish pigment cell types (xanthophore (orange), melanocyte (black) and iridophore (blue)) are generated from early neural crest cells (NCCs) by a mechanism involving production of intermediate progenitors that are fate-restricted, respectively, to two or three pigment cell options: melanoiridoblasts (blue, black), generating melanocytes and iridophores only, and chromatoblasts, generating all pigment cell fates (black, blue, orange); other progenitors give rise to other derivatives (e.g., neuroglioblasts (cyan, green) to peripheral ganglia). Key molecular markers as revealed by in situ hybridization are indicated in italics for each proposed cell type; note how recent studies have shown that maintenance and upregulation (rather than de novo transcription) of expression of these markers often characterizes the fate specification and differentiation phases, as indicated here by number of ‘+’ symbols after each gene name. In this diagram, the absence of a marker (e.g., the neuronal transcription factors neurog1 and phox2bb) is significant. Key transcription factor genes (yellow) and environmental signaling molecules (red) driving specific fate choices are indicated on the transition arrows. Note that the development of xanthophores has been less well studied. The mechanisms driving the formation of each intermediate progenitor have not been proposed.
More directly, the recent attempt to test the PFR model for pigment cell development has refuted the idea. Researchers used sensitive detection of key fate-specific gene expression using Nanostring technology to explore the transcriptome of dissociated zebrafish NCCs throughout the embryonic and very early larval periods, supplemented by both single cell TaqMan assays and sensitive in situ hybridization by RNAscope [81]. Neural crest-derived cells were isolated using fluorescence-activated cell sorting following a transgenic labelling approach. Researchers used a well-characterized sox10:Cre driver line [82] to label all neural crest-derived cells after their expression of sox10. Nanostring profiling of marker genes was focused on those genes known to have key roles in pigment cell development, especially melanocytes and iridophores, to ensure sensitivity to detection of the proposed intermediates in pigment cell development, but also included selected such markers for other NCC fates; the single cell Taqman assays assessed complementary markers for neuronal fates. Current study revealed the unexpected expression of markers of non-pigment cell fates (including, strikingly, neuronal markers neurog1 and phox2b) overlapping with those of all pigment cell fates; this was even true of control melanocyte and xanthophore cells isolated from 72 hpf larvae. It is currently unclear whether these contrasting observations stem from the increased sensitivity afforded by the Nanostring approach compared to scRNA-seq, or to more extensive disaggregation process leading to cells experiencing a more neutral environment allowing ‘relaxation’ of environmental signal-dependent fate specification, or a mixture of both; however, the co-expression of phox2b in many premigratory and a subset of postmigratory cells in glial locations strongly supports the increased sensitivity hypothesis [81], leading us to conclude that these cells are not partially restricted. Hence, researchers reinterpret the apparent partial restriction seen in ISH studies as partially reflecting technical limitations (most studies only look at one or two markers) and observer bias (where more markers are examined, lower level signals are often dismissed), but also reflecting their environmentally-induced bias towards subsets of fates, rather than the absolute fate restriction. Note that this conclusion would seem equally applicable to neural progenitors as to pigment cell progenitors. Indeed, transgenic fate mapping of cells expressing leukocyte tyrosine kinase (ltk), a marker of premigratory NCCs and of the iridophore lineage, encoding a receptor tyrosine kinase that is crucial for iridophore fate specification and proposed as a marker of putative chromatoblasts in vivo [39], reveals that early ltk expressing cells show multipotency, for all pigment cell fates, but strikingly also for neural fates, provides independent support for the conclusion that zebrafish pigment cell progenitors have unexpectedly broad multipotency [81]. Finally, in some cases, researchers were able to show by RNAscope ISH that post-migratory NCCs showed detectable expression of unexpected combinations of markers, with Schwann cell precursors on the posterior lateral line (sensory) nerve having co-expression of phox2b (autonomic neuron) and ltk (iridophore) markers [81]. Furthermore, researchers note that Current data provided no evidence for bipotent melanoiridoblasts either, despite the fact that Current selected gene set was highly enriched for early melanocyte and iridophore markers. None of these observations are consistent with the PFR model.
A number of recent studies have documented scRNA-seq profiles from zebrafish NC-derived cells [60][79][80]. In general, these have confirmed observations made by standard ISH that premigratory NCCs show early expression of pigment cell markers, indicating early fate specification of these lineages. For example, a recent characterization of 24 hpf trunk NCCs highlighted expression of xanthophore markers [60], confirming old observations of readily-detectable expression in premigratory and migratory trunk NCCs for multiple xanthophore genes; it is worth noting that whilst these cells are likely premigratory, their location adjacent to the epidermis dorsal to the neural tube is also a postmigratory location for xanthophores and melanocytes, so that early differentiation in this region may not be that surprising. However, it has become clear in recent years that these cells, and also migrating NCCs, are often specified to >1 fate, and the scRNA-seq data imply that they may be expressing markers of 2 fates at significant levels [60][79][80]. Detection of such a signature is very much limited by technical sensitivity to low-level expression in these studies, although that is rarely commented upon. The identified cell clusters also show low-level expression (defined as a low proportion of cells showing positive expression) of key markers of other fates too, hinting at the possibility that these cells retain very broad multipotency; thus, the difference between the scRNA-seq and Nanostring data is most likely one of sensitivity of detection of low-level expression, rather than fundamental differences in the biology observed. Indeed, sensitivity of detection appears to be an issue, since the 24 hpf scRNA-seq dataset does not identify ltk expression in the putative pigment cell progenitors, despite the fact that this has been repeatedly reported by whole-mount ISH [39][40][41]. This conclusion is reinforced by Current observations using an optimized RNAscope approach, where researchers see distinct, but low level, expression of phox2bb, a classic sympathetic/enteric neuron fate specification factor, in unexpected locations, including in numerous glial cells, likely Schwann cell precursors, even in 72 hpf larvae [81]. Researchers would argue that this expression clearly indicates the potential of these cells to adopt one of these sympathetic and enteric neuronal fates, regardless of whether this is realized in vivo. (A similar situation is observed for pigment cell markers—although the authors do not comment on this, their quantitative Hybridization Chain Reaction ISH shows wide heterogeneity of expression levels of xanthophore markers aox5, slc2a15b, and gjb8, with some cells in their figures clearly showing just a handful of mRNAs [60]). Researchers note too that work in mouse and lamprey has recently provided evidence for a contribution of Schwann cell precursors to the enteric neuron population [83][84]; Researchers would predict that the same is likely in zebrafish. Finally, researchers note how Current observations support the growing awareness of the breadth of multipotency of Schwann cell precursors [31][85][86][87][88][89].
3.3. Broad Multipotency of Melanocyte Stem Cells
In mammals, hair pigmentation is derived from melanocytes within the hair bulb, with melanosomes being transferred to the keratinocytes that generate the hair shaft (reviewed in [47][90][91][92]). At each hair molt cycle, the hair shaft is regenerated from new keratinocytes derived from keratinocyte stem cells, and melanin is supplied de novo by new melanocytes generated from melanocyte stem cells (MSCs), retained in the hair bulge. In addition, it has been shown in mammals that cells with the stem cell properties of multipotency and self-renewal and capable of forming neurospheres, can be isolated from diverse embryonic (including the original neural crest stem cells (NCSCs) isolated by the Anderson lab from rodent NC) and adult locations including the skin and the gut (reviewed in [46][93][94][95]). Although sharing the properties noted, these NCSCs show differences in their apparent fate biases, reflecting their source location (e.g., [96]). MSCs and NCSCs have been shown to be NC derived; MSCs are probably best thought of as another, perhaps highly specialized, type of NCSC, since they have been shown to be multipotent too [48].
Fish are characterized by distinct embryonic/early larval and adult pigment patterns, with the former widely conserved and likely involved in camouflage and protection of the germline against UV damage, and the latter highly divergent, and important for diverse aspects of adult biology [97][98][99][100][101][102]. Early studies in zebrafish revealed that ablation of pigment cells in the early larva leads to their regeneration, apparently dependent upon a cryptic source of pigment cell precursors [103][104]. Subsequently it was shown that the majority of adult pigment cells are generated de novo during metamorphosis, with the likely exception of an unknown proportion of the adult xanthophores which result from a dedifferentiation-proliferation-differentiation process of the embryonically-derived skin xanthophores [105][106][107][108][109][110]. The source of the de novo pigment cells is hypothesized to be a progenitor cell, likely a stem cell, equivalent to the MSC, and initially named as such. Early studies showed that these cells are NC derived, and that ErbB signaling plays a key role in their being set aside as a quiescent source of pigment cells [105].
However, the identity, number and location of the progenitor cells remain unclear. Pioneering studies indicated that the stem cells are either associated with the peripheral nervous system and/or use the peripheral nerves to deliver their progeny to the skin [111]. A set of cells within the dorsal root ganglia marked with an mitfa:gfp transgene became the first well-defined source, and were named MSCs [112], but subsequent work indicates there may well be other such sources associated with other parts of the PNS [113]; indeed it may be that Schwann cell precursors throughout the PNS are (or include) these stem cells [85][111][114][115]. Study of the MSCs soon showed that, in addition to melanocytes, they generate xanthophores and iridophores, but also neurons and glia of the PNS [116]. Thus, they appear to be broadly multipotent, and hence equivalent to the diverse NC-derived neural crest stem cells (NCSCs) identified in mammals, although their strict fulfilment of the stem cell property of self-renewal has not been formally tested. From Current perspective here, the key observation is the retention even in the adult of a subset of NC-derived cells that generally sit in a quiescent state, maintained by the local niche, but that retain a clear multipotency when activated, whether at metamorphosis, during regeneration or in tissue maintenance.
References
- Le Douarin, N.M.; Kalcheim, C. The Neural Crest, 2nd ed.; Cambridge University Press: Cambridge, UK, 1999.
- Dupin, E.; Baroffio, A.; Dulac, C.; Cameron-Curry, P.; Le Douarin, N. Schwann-cell differentiation in clonal cultures of the neural crest, as evidenced by the anti-Schwann cell myelin protein monoclonal antibody. Proc. Natl. Acad. Sci. USA 1990, 87, 1119–1123.
- Sieber-Blum, M.; Cohen, A.M. Clonal analysis of quail neural crest cells: They are pluripotent and differentiate in vitro in the absence of noncrest cells. Dev. Biol. 1980, 80, 96–106.
- Sieber-Blum, M. Commitment of neural crest cells to the sensory neuron lineage. Science 1989, 243, 1608–1611.
- Ito, K.; Morita, T.; Sieber-Blum, M. In vitro clonal analysis of mouse neural crest development. Dev. Biol. 1993, 157, 517–525.
- Ito, K.; Sieber-Blum, M. Pluripotent and developmentally restricted neural-crest-derived cells in posterior visceral arches. Dev. Biol. 1993, 156, 191–200.
- Le Douarin, N.M. Cell line segregation during peripheral nervous system ontogeny. Science 1986, 231, 1515–1522.
- Baroffio, A.; Dupin, E.; Le Douarin, N. Clone-forming ability and differentiation potential of migratory neural crest cells. Proc. Natl. Acad. Sci. USA 1988, 85, 5325–5329.
- Baroffio, A.; Dupin, E.; Le Douarin, N.M. Common precursors for neural and mesectodermal derivatives in the cephalic neural crest. Development 1991, 112, 301–305.
- Weston, J.A. Neural crest cell development. Prog. Clin. Biol. Res. 1982, 85 Pt B, 359–379.
- Weston, J.A. Regulation of neural crest cell migration and differentiation. In Cell Interactions and Development: Molecular Mechanisms; Yamada, K.M., Ed.; John Wiley and Sons, Inc: Hoboken, NJ, USA, 1983; pp. 153–184.
- Weston, J.A. Sequential Segregation and Fate of Developmentally Restricted Intermediate Cell Populations in the Neural Crest Lineage. Curr. Topics Dev. Biol. 1991, 25, 133–153.
- Bronner-Fraser, M.; Fraser, S.E. Cell lineage analysis reveals multipotency of some avian neural crest cells. Nature 1988, 335, 161–164.
- Bronner-Fraser, M.; Fraser, S. Developmental potential of avian trunk neural crest cells in situ. Neuron 1989, 3, 755–766.
- Fraser, S.E.; Bronner-Fraser, M. Migrating neural crest cells in the trunk of the avian embryo are multipotent. Development 1991, 112, 913–920.
- Hari, L.; Miescher, I.; Shakhova, O.; Suter, U.; Chin, L.; Taketo, M.; Richardson, W.D.; Kessaris, N.; Sommer, L. Temporal control of neural crest lineage generation by Wnt/beta-catenin signaling. Development 2012, 139, 2107–2117.
- Stemple, D.L.; Anderson, D.J. Isolation of a stem cell for neurons and glia from the mammalian neural crest. Cell 1992, 71, 973–985.
- Shah, N.M.; Marchionni, M.A.; Isaacs, I.; Stroobant, P.; Anderson, D.J. Glial growth-factor restricts mammalian neural crest stem-cells to a glial fate. Cell 1994, 77, 349–360.
- Shah, N.M.; Groves, A.K.; Anderson, D.J. Alternative neural crest cell fates are instructively promoted by TGFbeta superfamily members. Cell 1996, 85, 331–343.
- Lo, L.; Sommer, L.; Anderson, D.J. MASH1 maintains competence for BMP2-induced neuronal differentiation in post-migratory neural crest cells. Curr. Biol. 1997, 7, 440–450.
- Shah, N.M.; Anderson, D.J. Integration of multiple instructive cues by neural crest stem cells reveals cell-intrinsic biases in relative growth factor responsiveness. Proc. Natl. Acad. Sci. USA 1997, 94, 11369–11374.
- Perez, S.E.; Rebelo, S.; Anderson, D.J. Early specification of sensory neuron fate revealed by expression and function of neurogenins in the chick embryo. Development 1999, 126, 1715–1728.
- Kim, J.; Lo, L.; Dormand, E.; Anderson, D.J. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron 2003, 38, 17–31.
- Morrison, S.J.; Perez, S.E.; Qiao, Z.; Verdi, J.M.; Hicks, C.; Weinmaster, G.; Anderson, D.J. Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell 2000, 101, 499–510.
- Reissmann, E.; Ernsberger, U.; Francis-West, P.; Rueger, D.; Brickell, P.; Rohrer, H. Involvement of bone morphogenetic protein-4 and bone morphogenetic protein-7 in the differentiation of the adrenergic phenotype in developing sympathetic neurons. Development 1996, 122, 2079–2088.
- Li, Y.; Vieceli, F.M.; Gonzalez, W.G.; Li, A.; Tang, W.; Lois, C.; Bronner, M.E. In Vivo Quantitative Imaging Provides Insights into Trunk Neural Crest Migration. Cell Rep. 2019, 26, 1489–1500.e1483.
- Erickson, C.A.; Duong, T.D.; Tosney, K.W. Descriptive and experimental analysis of the dispersion of neural crest cells along the dorsolateral path and their entry into ectoderm in the chick embryo. Dev. Biol. 1992, 151, 251–272.
- Reedy, M.V.; Faraco, C.D.; Erickson, C.A. The delayed entry of thoracic neural crest cells into the dorsolateral path is a consequence of the late emigration of melanogenic neural crest cells from the neural tube. Dev. Biol. 1998, 200, 234–246.
- Krispin, S.; Nitzan, E.; Kassem, Y.; Kalcheim, C. Evidence for a dynamic spatiotemporal fate map and early fate restrictions of premigratory avian neural crest. Development 2010, 137, 585–595.
- Nitzan, E.; Krispin, S.; Pfaltzgraff, E.R.; Klar, A.; Labosky, P.A.; Kalcheim, C. A dynamic code of dorsal neural tube genes regulates the segregation between neurogenic and melanogenic neural crest cells. Development 2013, 140, 2269–2279.
- Nitzan, E.; Pfaltzgraff, E.R.; Labosky, P.A.; Kalcheim, C. Neural crest and Schwann cell progenitor-derived melanocytes are two spatially segregated populations similarly regulated by Foxd3. Proc. Natl. Acad. Sci. USA 2013, 110, 12709–12714.
- Kitamura, K.; Takiguchi-Hayashi, K.; Sezaki, M.; Yamamoto, H.; Takeuchi, T. Avian neural crest cells express a melanogenic trait migration from the neural tube: Observations with the antibody, “MEBL-1”. Development 1992, 114, 367–378.
- Serbedzija, G.N.; Bronner-Fraser, M.; Fraser, S.E. A vital dye analysis of the timing and pathways of avian trunk neural crest cell migration. Development 1989, 106, 809–816.
- Gilbert, S.F.; Barresi, M.J.F. Developmental Biology, 11th ed.; Sinauer Associates Inc.: Sunderland, MD, USA, 2016.
- Baggiolini, A.; Varum, S.; Mateos, J.M.; Bettosini, D.; John, N.; Bonalli, M.; Ziegler, U.; Dimou, L.; Clevers, H.; Furrer, R.; et al. Premigratory and migratory neural crest cells are multipotent in vivo. Cell Stem Cell 2015, 16, 314–322.
- Soldatov, R.; Kaucka, M.; Kastriti, M.E.; Petersen, J.; Chontorotzea, T.; Englmaier, L.; Akkuratova, N.; Yang, Y.; Haring, M.; Dyachuk, V.; et al. Spatiotemporal structure of cell fate decisions in murine neural crest. Science 2019, 364.
- Ling, I.T.C.; Sauka-Spengler, T. Early chromatin shaping predetermines multipotent vagal neural crest into neural, neuronal and mesenchymal lineages. Nat. Cell Biol. 2019, 21, 1504–1517.
- Kelsh, R.N.; Schmid, B.; Eisen, J.S. Genetic analysis of melanophore development in zebrafish embryos. Dev. Biol. 2000, 225, 277–293.
- Lopes, S.S.; Yang, X.; Muller, J.; Carney, T.J.; McAdow, A.R.; Rauch, G.J.; Jacoby, A.S.; Hurst, L.D.; Delfino-Machin, M.; Haffter, P.; et al. Leukocyte tyrosine kinase functions in pigment cell development. PLoS Genet. 2008, 4, e1000026.
- Petratou, K.; Subkhankulova, T.; Lister, J.A.; Rocco, A.; Schwetlick, H.; Kelsh, R.N. A systems biology approach uncovers the core gene regulatory network governing iridophore fate choice from the neural crest. PLoS Genet. 2018, 14, e1007402.
- Petratou, K.; Spencer, S.A.; Kelsh, R.N.; Lister, J.A. The MITF paralog tfec is required in neural crest development for fate specification of the iridophore lineage from a multipotent pigment cell progenitor. PLoS ONE 2021, 16, e0244794.
- Parichy, D.M.; Ransom, D.G.; Paw, B.; Zon, L.I.; Johnson, S.L. An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a subpopulation of adult melanocytes in the zebrafish, Danio rerio. Development 2000, 127, 3031–3044.
- Parichy, D.M.; Rawls, J.F.; Pratt, S.J.; Whitfield, T.T.; Johnson, S.L. Zebrafish sparse corresponds to an orthologue of c-kit and is required for the morphogenesis of a subpopulation of melanocytes, but is not essential for hematopoiesis or primordial germ cell development. Development 1999, 126, 3425–3436.
- Curran, K.; Raible, D.W.; Lister, J.A. Foxd3 controls melanophore specification in the zebrafish neural crest by regulation of Mitf. Dev. Biol. 2009, 332, 408–417.
- Lister, J.A.; Robertson, C.P.; Lepage, T.; Johnson, S.L.; Raible, D.W. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development 1999, 126, 3757–3767.
- Delfino-Machin, M.; Chipperfield, T.R.; Rodrigues, F.S.; Kelsh, R.N. The proliferating field of neural crest stem cells. Dev. Dyn. 2007, 236, 3242–3254.
- Nishimura, E.K. Melanocyte stem cells: A melanocyte reservoir in hair follicles for hair and skin pigmentation. Pigment Cell Melanoma Res. 2011, 24, 401–410.
- Watanabe, N.; Motohashi, T.; Nishioka, M.; Kawamura, N.; Hirobe, T.; Kunisada, T. Multipotency of melanoblasts isolated from murine skin depends on the Notch signal. Dev. Dyn. 2016, 245, 460–471.
- McGraw, H.F.; Nechiporuk, A.; Raible, D.W. Zebrafish dorsal root ganglia neural precursor cells adopt a glial fate in the absence of neurogenin1. J. Neurosci. 2008, 28, 12558–12569.
- Carney, T.J.; Dutton, K.A.; Greenhill, E.; Delfino-Machin, M.; Dufourcq, P.; Blader, P.; Kelsh, R.N. A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development 2006, 133, 4619–4630.
- Pei, D.; Luther, W.; Wang, W.; Paw, B.H.; Stewart, R.A.; George, R.E. Distinct neuroblastoma-associated alterations of PHOX2B impair sympathetic neuronal differentiation in zebrafish models. PLoS Genet. 2013, 9, e1003533.
- Elworthy, S.; Pinto, J.P.; Pettifer, A.; Cancela, M.L.; Kelsh, R.N. Phox2b function in the enteric nervous system is conserved in zebrafish and is sox10-dependent. Mech. Dev. 2005, 122, 659–669.
- Dutton, K.A.; Pauliny, A.; Lopes, S.S.; Elworthy, S.; Carney, T.J.; Rauch, J.; Geisler, R.; Haffter, P.; Kelsh, R.N. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development 2001, 128, 4113–4125.
- Delfino-Machin, M.; Madelaine, R.; Busolin, G.; Nikaido, M.; Colanesi, S.; Camargo-Sosa, K.; Law, E.W.; Toppo, S.; Blader, P.; Tiso, N.; et al. Sox10 contributes to the balance of fate choice in dorsal root ganglion progenitors. PLoS ONE 2017, 12, e0172947.
- Budi, E.H.; Patterson, L.B.; Parichy, D.M. Embryonic requirements for ErbB signaling in neural crest development and adult pigment pattern formation. Development 2008, 135, 2603–2614.
- Honjo, Y.; Kniss, J.; Eisen, J.S. Neuregulin-mediated ErbB3 signaling is required for formation of zebrafish dorsal root ganglion neurons. Development 2008, 135, 2615–2625.
- Haffter, P.; Odenthal, J.; Mullins, M.C.; Lin, S.; Farrell, M.J.; Vogelsang, E.; Haas, F.; Brand, M.; van Eeden, F.J.; Furutani-Seiki, M.; et al. Mutations affecting pigmentation and shape of the adult zebrafish. Dev. Genes Evol. 1996, 206, 260–276.
- Kelsh, R.N.; Brand, M.; Jiang, Y.J.; Heisenberg, C.P.; Lin, S.; Haffter, P.; Odenthal, J.; Mullins, M.C.; van Eeden, F.J.; Furutani-Seiki, M.; et al. Zebrafish pigmentation mutations and the processes of neural crest development. Development 1996, 123, 369–389.
- Odenthal, J.; Rossnagel, K.; Haffter, P.; Kelsh, R.; Vogelsang, E.; Brand, M.; van Eeden, F.; Furutani-Seiki, M.; Granato, M.; Hammerschmidt, M.; et al. Mutations affecting xanthophore pigmentation in the zebrafish, Danio rerio. Development 1996, 123, 391–398.
- Lencer, E.; Prekeris, R.; Artinger, K.B. Single-cell RNA analysis identifies pre-migratory neural crest cells expressing markers of differentiated derivatives. Elife 2021, 10, e66078.
- Johnson, S.L.; Africa, D.; Walker, C.; Weston, J.A. Genetic control of adult pigment stripe development in zebrafish. Dev. Biol. 1995, 167, 27–33.
- Parichy, D.M.; Mellgren, E.M.; Rawls, J.F.; Lopes, S.S.; Kelsh, R.N.; Johnson, S.L. Mutational analysis of endothelin receptor b1 (rose) during neural crest and pigment pattern development in the zebrafish Danio rerio. Dev. Biol. 2000, 227, 294–306.
- Rawls, J.F.; Johnson, S.L. Zebrafish kit mutation reveals primary and secondary regulation of melanocyte development during fin stripe regeneration. Development 2000, 127, 3715–3724.
- Johnson, S.L.; Nguyen, A.N.; Lister, J.A. Mitfa is required at multiple stages of melanocyte differentiation but not to establish the melanocyte stem cell. Dev. Biol. 2011, 350, 405–413.
- Elworthy, S.; Lister, J.A.; Carney, T.J.; Raible, D.W.; Kelsh, R.N. Transcriptional regulation of mitfa accounts for the sox10 requirement in zebrafish melanophore development. Development 2003, 130, 2809–2818.
- Schartl, M.; Larue, L.; Goda, M.; Bosenberg, M.W.; Hashimoto, H.; Kelsh, R.N. What is a vertebrate pigment cell? Pigment Cell Melanoma Res. 2016, 29, 8–14.
- Raible, D.W.; Eisen, J.S. Restriction of neural crest cell fate in the trunk of the embryonic zebrafish. Development 1994, 120, 495–503.
- Schilling, T.F.; Kimmel, C.B. Segment and cell type lineage restrictions during pharyngeal arch development in the zebrafish embryo. Development 1994, 120, 483–494.
- Minchin, J.E.; Hughes, S.M. Sequential actions of Pax3 and Pax7 drive xanthophore development in zebrafish neural crest. Dev. Biol. 2008, 317, 508–522.
- Curran, K.; Lister, J.A.; Kunkel, G.R.; Prendergast, A.; Parichy, D.M.; Raible, D.W. Interplay between Foxd3 and Mitf regulates cell fate plasticity in the zebrafish neural crest. Dev. Biol. 2010, 344, 107–118.
- Krauss, J.; Frohnhofer, H.G.; Walderich, B.; Maischein, H.M.; Weiler, C.; Irion, U.; Nusslein-Volhard, C. Endothelin signalling in iridophore development and stripe pattern formation of zebrafish. Biol. Open 2014, 3, 503–509.
- Fadeev, A.; Krauss, J.; Singh, A.P.; Nusslein-Volhard, C. Zebrafish Leucocyte tyrosine kinase controls iridophore establishment, proliferation and survival. Pigment Cell Melanoma Res. 2016, 29, 284–296.
- Nord, H.; Dennhag, N.; Muck, J.; von Hofsten, J. Pax7 is required for establishment of the xanthophore lineage in zebrafish embryos. Mol. Biol. Cell 2016, 27, 1853–1862.
- Fadeev, A.; Mendoza-Garcia, P.; Irion, U.; Guan, J.; Pfeifer, K.; Wiessner, S.; Serluca, F.; Singh, A.P.; Nusslein-Volhard, C.; Palmer, R.H. ALKALs are in vivo ligands for ALK family receptor tyrosine kinases in the neural crest and derived cells. Proc. Natl. Acad. Sci. USA 2018, 115, E630–E638.
- Bagnara, J.T.; Matsumoto, J.; Ferris, W.; Frost, S.K.; Turner, W.A., Jr.; Tchen, T.T.; Taylor, J.D. Common origin of pigment cells. Science 1979, 203, 410–415.
- Kelsh, R.N.; Inoue, C.; Momoi, A.; Kondoh, H.; Furutani-Seiki, M.; Ozato, K.; Wakamatsu, Y. The Tomita collection of medaka pigmentation mutants as a resource for understanding neural crest cell development. Mech. Dev. 2004, 121, 841–859.
- Kimura, T.; Nagao, Y.; Hashimoto, H.; Yamamoto-Shiraishi, Y.; Yamamoto, S.; Yabe, T.; Takada, S.; Kinoshita, M.; Kuroiwa, A.; Naruse, K. Leucophores are similar to xanthophores in their specification and differentiation processes in medaka. Proc. Natl. Acad. Sci. USA 2014, 111, 7343–7348.
- Nagao, Y.; Suzuki, T.; Shimizu, A.; Kimura, T.; Seki, R.; Adachi, T.; Inoue, C.; Omae, Y.; Kamei, Y.; Hara, I.; et al. Sox5 functions as a fate switch in medaka pigment cell development. PLoS Genet. 2014, 10, e1004246.
- Saunders, L.M.; Mishra, A.K.; Aman, A.J.; Lewis, V.M.; Toomey, M.B.; Packer, J.S.; Qiu, X.; McFaline-Figueroa, J.L.; Corbo, J.C.; Trapnell, C.; et al. Thyroid hormone regulates distinct paths to maturation in pigment cell lineages. Elife 2019, 8, e45181.
- Howard, A.G.T.; Baker, P.A.; Ibarra-Garcia-Padilla, R.; Moore, J.A.; Rivas, L.J.; Tallman, J.J.; Singleton, E.W.; Westheimer, J.L.; Corteguera, J.A.; Uribe, R.A. An atlas of neural crest lineages along the posterior developing zebrafish at single-cell resolution. Elife 2021, 10, e60005.
- Nikaido, M.; Subkhankulova, T.; Kasianov, A.; Uroshlev, L.; Camargo Sosa, K.; Bavister, G.; Yang, X.; Rodrigues, F.S.L.M.; Carney, T.J.; Dawes, J.H.P.; et al. Zebrafish pigment cells develop directly from highly multipotent progenitors. bioRxiv 2021.
- Rodrigues, F.S.; Doughton, G.; Yang, B.; Kelsh, R.N. A novel transgenic line using the Cre-lox system to allow permanent lineage-labeling of the zebrafish neural crest. Genesis 2012, 50, 750–757.
- Green, S.A.; Uy, B.R.; Bronner, M.E. Ancient evolutionary origin of vertebrate enteric neurons from trunk-derived neural crest. Nature 2017, 544, 88–91.
- Espinosa-Medina, I.; Jevans, B.; Boismoreau, F.; Chettouh, Z.; Enomoto, H.; Muller, T.; Birchmeier, C.; Burns, A.J.; Brunet, J.F. Dual origin of enteric neurons in vagal Schwann cell precursors and the sympathetic neural crest. Proc. Natl. Acad. Sci. USA 2017, 114, 11980–11985.
- Adameyko, I.; Lallemend, F.; Aquino, J.B.; Pereira, J.A.; Topilko, P.; Muller, T.; Fritz, N.; Beljajeva, A.; Mochii, M.; Liste, I.; et al. Schwann cell precursors from nerve innervation are a cellular origin of melanocytes in skin. Cell 2009, 139, 366–379.
- Adameyko, I.; Lallemend, F.; Furlan, A.; Zinin, N.; Aranda, S.; Kitambi, S.S.; Blanchart, A.; Favaro, R.; Nicolis, S.; Lubke, M.; et al. Sox2 and Mitf cross-regulatory interactions consolidate progenitor and melanocyte lineages in the cranial neural crest. Development 2012, 139, 397–410.
- Furlan, A.; Adameyko, I. Schwann cell precursor: A neural crest cell in disguise? Dev. Biol. 2018, 444, S25–S35.
- Furlan, A.; Dyachuk, V.; Kastriti, M.E.; Calvo-Enrique, L.; Abdo, H.; Hadjab, S.; Chontorotzea, T.; Akkuratova, N.; Usoskin, D.; Kamenev, D.; et al. Multipotent peripheral glial cells generate neuroendocrine cells of the adrenal medulla. Science 2017, 357.
- Solovieva, T.; Bronner, M. Schwann cell precursors: Where they come from and where they go. Cells Dev. 2021, 166, 203686.
- Li, H.; Hou, L. Regulation of melanocyte stem cell behavior by the niche microenvironment. Pigment Cell Melanoma Res. 2018, 31, 556–569.
- Qiu, W.; Chuong, C.M.; Lei, M. Regulation of melanocyte stem cells in the pigmentation of skin and its appendages: Biological patterning and therapeutic potentials. Exp. Dermatol. 2019, 28, 395–405.
- Arnheiter, H.; Debbache, J. Development of Melanin-Bearing Pigment Cells in Birds and Mammals. In Pigments, Pigment Cells and Pigment Patterns; Hashimoto, H., Goda, M., Futahashi, R., Kelsh, R., Akiyama, T., Eds.; Springer Nature Singapore Pte Ltd.: Singapore, 2021; pp. 185–208.
- Sieber-Blum, M. Human epidermal neural crest stem cells as candidates for cell-based therapies, disease modeling, and drug discovery. Birth Defects Res. C Embryo Today 2014, 102, 221–226.
- Liu, J.A.; Cheung, M. Neural crest stem cells and their potential therapeutic applications. Dev. Biol. 2016, 419, 199–216.
- Moghadasi Boroujeni, S.; Koontz, A.; Tseropoulos, G.; Kerosuo, L.; Mehrotra, P.; Bajpai, V.K.; Selvam, S.R.; Lei, P.; Bronner, M.E.; Andreadis, S.T. Neural crest stem cells from human epidermis of aged donors maintain their multipotency in vitro and in vivo. Sci. Rep. 2019, 9, 9750.
- White, P.M.; Morrison, S.J.; Orimoto, K.; Kubu, C.J.; Verdi, J.M.; Anderson, D.J. Neural crest stem cells undergo cell-intrinsic developmental changes in sensitivity to instructive differentiation signals. Neuron 2001, 29, 57–71.
- Parichy, D.M.; Liang, Y. Evolution of Pigment Patterning in Teleosts. In Pigments, Pigment Cells and Pigment Patterns; Hashimoto, H., Goda, M., Futahashi, R., Kelsh, R., Akiyama, T., Eds.; Springer Nature Singapore Pte Ltd.: Singapore, 2021; pp. 309–342.
- Owen, J.; Yates, C.; Kelsh, R.N. Pigment Patterning in Teleosts. In Pigments, Pigment Cells and Pigment Patterns; Hashimoto, H., Goda, M., Futahashi, R., Kelsh, R., Akiyama, T., Eds.; Springer Nature Singapore Pte Ltd.: Singapore, 2021; pp. 247–292.
- Parichy, D.M. Evolution of pigment cells and patterns: Recent insights from teleost fishes. Curr. Opin. Genet. Dev. 2021, 69, 88–96.
- Parichy, D.M. Advancing biology through a deeper understanding of zebrafish ecology and evolution. Elife 2015, 4, e05635.
- Engeszer, R.E.; Barbiano, L.A.; Ryan, M.J.; Parichy, D.M. Timing and plasticity of shoaling behaviour in the zebrafish, Danio rerio. Anim. Behav. 2007, 74, 1269–1275.
- Irion, U.; Singh, A.P.; Nusslein-Volhard, C. The Developmental Genetics of Vertebrate Color Pattern Formation: Lessons from Zebrafish. Curr. Top. Dev. Biol. 2016, 117, 141–169.
- Yang, C.T.; Sengelmann, R.D.; Johnson, S.L. Larval melanocyte regeneration following laser ablation in zebrafish. J. Investig. Dermatol. 2004, 123, 924–929.
- Yang, C.T.; Johnson, S.L. Small molecule-induced ablation and subsequent regeneration of larval zebrafish melanocytes. Development 2006, 133, 3563–3573.
- Hultman, K.A.; Budi, E.H.; Teasley, D.C.; Gottlieb, A.Y.; Parichy, D.M.; Johnson, S.L. Defects in ErbB-dependent establishment of adult melanocyte stem cells reveal independent origins for embryonic and regeneration melanocytes. PLoS Genet. 2009, 5, e1000544.
- Hultman, K.A.; Johnson, S.L. Differential contribution of direct-developing and stem cell-derived melanocytes to the zebrafish larval pigment pattern. Dev. Biol. 2010, 337, 425–431.
- Tryon, R.C.; Higdon, C.W.; Johnson, S.L. Lineage relationship of direct-developing melanocytes and melanocyte stem cells in the zebrafish. PLoS ONE 2011, 6, e21010.
- Parichy, D.M.; Spiewak, J.E. Origins of adult pigmentation: Diversity in pigment stem cell lineages and implications for pattern evolution. Pigment Cell Melanoma Res. 2015, 28, 31–50.
- McMenamin, S.K.; Bain, E.J.; McCann, A.E.; Patterson, L.B.; Eom, D.S.; Waller, Z.P.; Hamill, J.C.; Kuhlman, J.A.; Eisen, J.S.; Parichy, D.M. Thyroid hormone-dependent adult pigment cell lineage and pattern in zebrafish. Science 2014, 345, 1358–1361.
- Mahalwar, P.; Walderich, B.; Singh, A.P.; Nusslein-Volhard, C. Local reorganization of xanthophores fine-tunes and colors the striped pattern of zebrafish. Science 2014, 345, 1362–1364.
- Budi, E.H.; Patterson, L.B.; Parichy, D.M. Post-embryonic nerve-associated precursors to adult pigment cells: Genetic requirements and dynamics of morphogenesis and differentiation. PLoS Genet. 2011, 7, e1002044.
- Dooley, C.M.; Mongera, A.; Walderich, B.; Nusslein-Volhard, C. On the embryonic origin of adult melanophores: The role of ErbB and Kit signalling in establishing melanophore stem cells in zebrafish. Development 2013, 140, 1003–1013.
- Camargo-Sosa, K.; Colanesi, S.; Muller, J.; Schulte-Merker, S.; Stemple, D.; Patton, E.E.; Kelsh, R.N. Endothelin receptor Aa regulates proliferation and differentiation of Erb-dependent pigment progenitors in zebrafish. PLoS Genet. 2019, 15, e1007941.
- Kelsh, R.N.; Barsh, G.S. A nervous origin for fish stripes. PLoS Genet. 2011, 7, e1002081.
- Adameyko, I.; Lallemend, F. Glial versus melanocyte cell fate choice: Schwann cell precursors as a cellular origin of melanocytes. Cell Mol. Life Sci. 2010, 67, 3037–3055.
- Singh, A.P.; Dinwiddie, A.; Mahalwar, P.; Schach, U.; Linker, C.; Irion, U.; Nusslein-Volhard, C. Pigment Cell Progenitors in Zebrafish Remain Multipotent through Metamorphosis. Dev. Cell 2016, 38, 316–330.
More
Information
Subjects:
Developmental Biology; Genetics & Heredity
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.0K
Revisions:
2 times
(View History)
Update Date:
27 Dec 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




