Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ivo Sauman | + 2951 word(s) | 2951 | 2021-12-23 03:48:34 | | | |
| 2 | Amina Yu | -2 word(s) | 2949 | 2021-12-24 02:17:41 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Sauman, I. Filippi’s Glands and Silk Moths Cocoon Construction. Encyclopedia. Available online: https://encyclopedia.pub/entry/17501 (accessed on 08 February 2026).
Sauman I. Filippi’s Glands and Silk Moths Cocoon Construction. Encyclopedia. Available at: https://encyclopedia.pub/entry/17501. Accessed February 08, 2026.
Sauman, Ivo. "Filippi’s Glands and Silk Moths Cocoon Construction" Encyclopedia, https://encyclopedia.pub/entry/17501 (accessed February 08, 2026).
Sauman, I. (2021, December 23). Filippi’s Glands and Silk Moths Cocoon Construction. In Encyclopedia. https://encyclopedia.pub/entry/17501
Sauman, Ivo. "Filippi’s Glands and Silk Moths Cocoon Construction." Encyclopedia. Web. 23 December, 2021.
Copy Citation
Filippi’s glands (FGs), formerly also called Lyonet’s glands, are accessory secretory structures of the labial (silk) glands of lepidopteran caterpillars, which were implicated to play an important role in the maturation of the silk material and the construction of the cocoon.
Filippi’s glands
Saturniidae
cocoon structure
Bombyx mori
silk
proteomic analysis
1. Silk Moth Species without Filippi’s Glands Construct Loose Cocoons
In previous study [1], it was identified six species of giant silkmoth (Saturniidae) that lack FGs throughout their larval development. Here it is interested whether the presence of FGs had effect on the type of cocoon constructed. It chose three representatives of the Saturniidae that lack FGs (Hyalophora cecropia, Samia cynthia, and Attacus atlas), two Saturniid species that have well-developed FGs (Antheraea polyphemus and Actias selene), and the silkworm B. mori from the sister family Bombycidae, which has well-developed FGs relative to the size of its spinneret. It was found that species without FGs spin loose cocoons in which the silk layers formed outer and inner envelopes with an intermediate silk scaffold (Figure 1), whereas species with well-developed FGs exclusively form compact cocoons with silk in a compact layer (Figure 2). Thus, the presence or absence of FGs was clearly correlated with the type of cocoon spun.
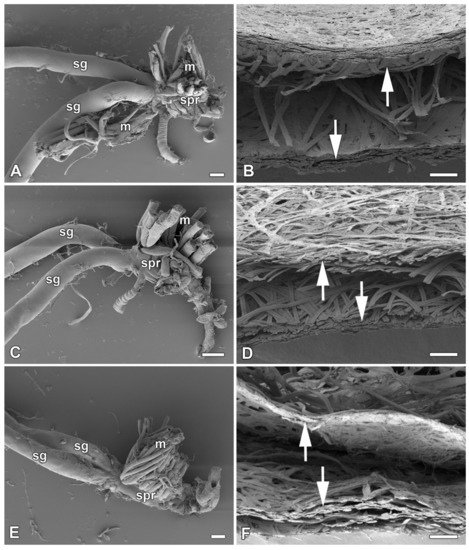
Figure 1. Spinning apparatus of last instar larvae without FGs and cross sections of their loose cocoons. (A,B) The cecropia silk moth, Hyalophora cecropia. (C,D) The ailanthus silk moth, Samia cynthia. (E,F) The Atlas moth, Attacus atlas. Scale bars: (A,C,E) = 100 µm; (B,D,F) = 200 µm. Abbreviations: silk glands (sg), silk press of the spinning apparatus (spr), muscles (m). The arrows depict two distinct layers of the cocoon.
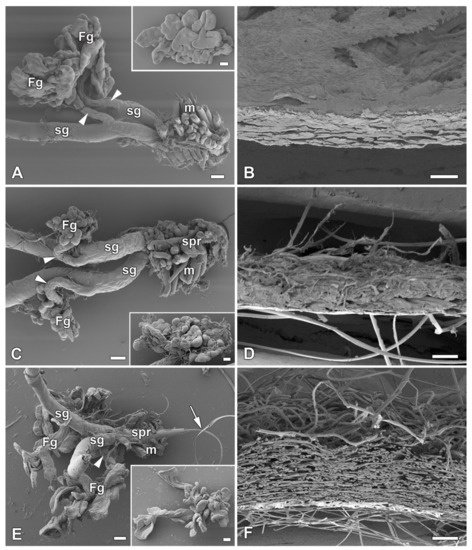
Figure 2. Spinning apparatus of last instar larvae with FGs and cross sections of their compact cocoons. (A,B) The Polyphemus moth, Antheraea polyphemus. (C,D) The Indian moon moth, Actias selene. (E,F) The commercial silkworm, Bombyx mori. Scale bars: (A,C,E) = 100 µm; (B,D,F) = 200 µm. Abbreviations: Filippi’s glands (Fg), silk glands (sg), silk press of the spinning apparatus (spr), muscles (m). The arrowheads indicate the ducts of the Filippi’s glands entering the lumen of the silk glands. The arrow points to the secreted silk fibre.
To further support this conclusion, it also compared all published results demonstrating the presence or absence of FGs in giant silkworms [2][1] with their respective cocoon architecture [3][4][5]. For the following species, Saturnia pavoniella, Antherina suraka, and Aglia tau, where the complete absence of FGs has been demonstrated [1] and whose cocoon structure has not yet been described in the literature, it used SEM to show that they form compact cocoons (Supplementary Materials Figure S1). The correlation between the presence of FGs and the type of cocoon architecture is shown in Figure 3. Larvae of Saturniid species with FGs exclusively construct the compact cocoons (Antheraea pernyi, Antheraea yamamai, Antheraea mylitta, A. polyphemus, A. selene, S. pavonia, S. pavoniella, Saturnia pyri, and A. suraka). In contrast, species of silk moths that do not have FGs spin loose cocoons (H. cecropia, C. promethea, S. cynthia, A. atlas). The most ancestral Saturniid species examined in this study, A. tau, has no FGs and forms a sparse silk cocoon (Supplementary Materials Figure S1C).
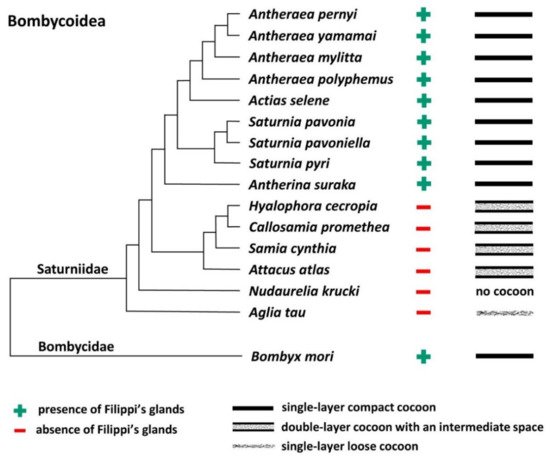
Figure 3. Phylogenetic tree of the superfamily Bombycoidea in relation to the presence/absence of FGs and the type of cocoon. The presence/absence of FGs is based on published data [2][1] and cocoon structure of A. mylitta, A. pernyi, A. yamamai, S. pavonia, S. pyri, and C. promethea was previously studied by Chen et al. [4]. The types of cocoons of the other species in this study are indicated in Figure 1 and Figure 2 and Figure S1 in the Supplementary Materials. The phylogenetic tree was based on Chen et al. [6] and Hamilton et al. [7].
2. Effect of Filippi’s Glands Removal in B. mori Larvae
To study the effect of FGs on cocoon architecture, it chose the silkworm B. mori, in which these glands are well developed and defined, and the larvae form compact cocoons. Before spinning begins, the larvae of the last instar empty their gut contents and then enter the wandering stage. After the onset of silk extrusion, it surgically removed the FGs and allowed the larvae to spin cocoons. The efficiency of cocoon construction after surgery was more than 61% (27 of 44 larvae) compared to control animals, where the efficiency was 75% (15/20). Removal of FGs had no obvious effect on larval spinning behavior. After resuming the spinning following the anesthesia and surgery (approximately 3–6 h), the time of cocoon construction was comparable to that of the non-operated control larvae (about 48 h).
However, when comparing the cocoon architecture between the operated and control animals, it was found a striking difference in the compactness and thickness of the cocoon wall structure. In the cocoons of the operated animals, the silk layers were loosely arranged compared to the very dense structure of the layers in the cocoons of the control animals (Figure 4). These differences were observed at the level of light microscopy (Figure 4A,B). Even more convincing evidence for these structural differences was provided by scanning electron microscopy (SEM) examination (Figure 4C,D). High-resolution SEM images of the outer and inner sides of the cocoon wall revealed striking differences in the arrangement of silk fibers. On both sides, cocoons from animals with extirpated FGs exhibited a much looser arrangement (Figure 4E–H). The most striking differences were observed on the inner side, where the cocoons of operated animals had a significantly lower coverage with sericins than the cocoons of control animals, where sericin covered a considerable part of the surface (Figure 4G,H).
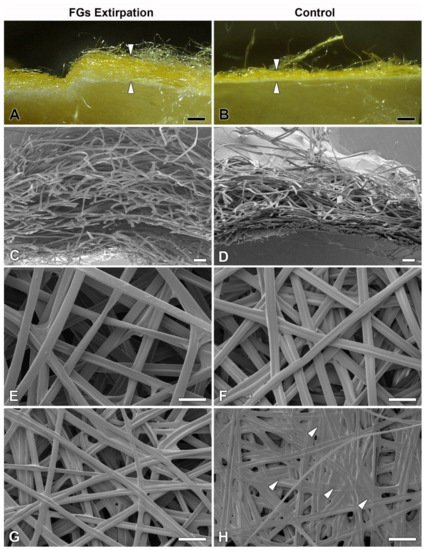
Figure 4. Effect of surgical removal of FGs in silkworm larvae on cocoon structure. (A) A macrophotographic image of the cross section of the cocoon wall spun by a B. mori larva after complete removal of both FGs. Note the much thicker and looser structure of the cocoon wall (arrowheads). (B) Corresponding image from a control animal with FGs intact. The arrowheads indicate the thickness of the normal compact cocoon wall. (C) A scanning electron micrograph showing a cross section of the architecture of the loose silk fibers of the cocoon wall of a B. mori larva with surgically removed FGs. (D) The silk fibers of a control larva with intact FGs are densely arranged and form a much more compact cocoon wall. (E,F) A microstructure of the cocoon silk fibers from the outside of the cocoon wall shows no visible differences between operated and control animals. (G,H) The inner sides of the cocoon walls show significant differences in the amount of a “glue-like” material deposited on the silk fibers of the control (arrowheads) compared to the operated larvae. Scale bars: (A,B) = 1 mm; (C,D) = 100 µm; (E–H) = 50 µm.
3. The Filippi’s Glands Have No Effect on pH Regulation or Lipid Synthesis
Because the FGs can be involved in the transport of ions [8], it investigated their possible role in regulating the pH of silk in ASGs, during spinning. To test this possibility, pH was measured in vivo using the indicator dye phenol red. No differences in silk pH or in the epithelial cells of the SGs were observed between the FGs extirpated and the control larvae. Significant differences in the pH of the different parts of the spinning apparatus were found only between the B. mori larvae that resumed silk extrusion after application of the phenol dye and those that did not. In both, operated and unoperated larvae, that did not resume silk spinning, the anterior part of the SGs contained clusters of cells with higher pH (approximately between 6.5 and 7.5) and silk-like material in the lumen of the SGs and FG ducts with pH values around 6.5 (Figure 5A,C). In contrast, larvae (both, operated and control) that had resumed silk extrusion had a much more acidic pH of the entire spinning apparatus including the silk material (pH below 5.0; Figure 5B,D).
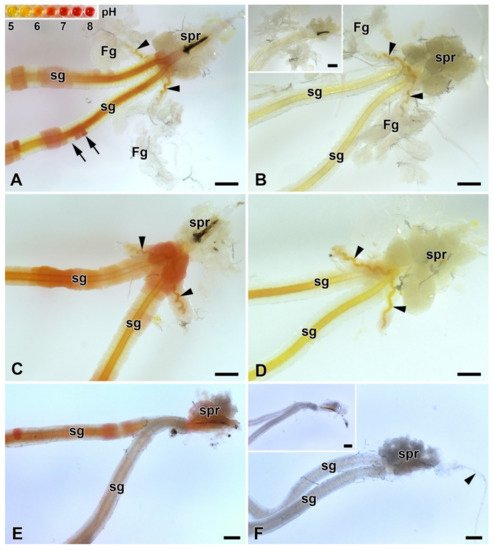
Figure 5. Acidification of the spinning apparatus of B. mori and the Eri silk moth (Samia cynthia) last instar larvae. (A) Brightfield micrograph of the intact spinning apparatus (control) of a “right now non-spinning” B. mori larva injected with the pH-sensitive dye (phenol red). The arrowheads show the ducts of the FGs entering the lumen of SGs. Inset: the pH scale of phenol red obtained with pH-calibrated solutions with 0.5 pH resolution (from pH 5 to 8). The arrows indicate significantly different pH values even in adjacent epithelial cells of the silk gland. (B) The spinning apparatus of a “right now spinning” B. mori larva injected with the phenol red dye. Arrowheads indicate the ducts of the FGs, which open into the lumen of sSGs. Inset: A spinning apparatus of Bombyx larva without phenol red injection (negative control for the effect of the phenol red injection). (C) Same as in (A) but in a larva that has had both FGs surgically removed. The arrowheads show the remains of the FGs ducts after their surgical removal. (D) The same as in (B) in a larva with extirpated FGs. The arrowheads show the residual ducts of the FGs opening into the lumen of the SGs. (E) Brightfield micrograph of the spinning apparatus of a “right now non-spinning” S. cynthia larva injected with the pH-sensitive dye phenol red. (F) The spinning apparatus of a “right now spinning” S. cynthia larva injected with the phenol red dye. Inset: a spinning apparatus of a Samia larva without phenol red injection (negative control for the effect of phenol red injection). All scale bars = 200 µm. Abbreviations: Filippi’s glands (Fg), silk (labial) glands (sg), silk press of the spinning apparatus (spr).
Similar changes in pH in ASGs between spinning and non-spinning larvae were also observed in a silk moth species lacking FGs (S. cynthia) when, after resumption of spinning, the pH values were lower than 5 and in those that did not start re-spinning after dye injection, the pH in some epithelial cells reached pH about 7.0 (Figure 5E,F).
Considering the published data demonstrating the presence of lipids in the FGs of A. mylitta [2] and active fatty acid biosynthesis in the FGs of B. mori [9], it examined the presence of lipids and fatty acids in FGs of B. mori, A. selene, and A. polyphemus. The presence of lipids in the FGs was not detected by Oil Red O staining in any of the species tested (Figure 6A,B; Supplementary Materials Figure S2A,B). Examination of the ultrastructure of FGs in B. mori also did not confirm the presence of lipid droplets or other vesicles with lubricating substances (Figure 6E,F). Since the presence of lipid droplets has been demonstrated in ASGs of some lepidopteran species [10][11], it also thoroughly analyzed this part of SGs. However, the presence of lipid components was not detected in the ASGs either, both in the above-mentioned species with FGs (Figure 6A,B,E,F; Supplementary Materials Figure S2A,B), and in species without FGs: H. cecropia and S. cynthia (Supplementary Materials Figure S2C–F).
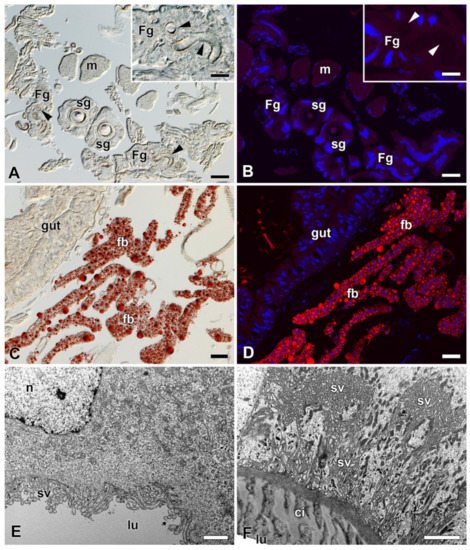
Figure 6. Detection of fat in the FGs and SGs of B. mori. (A) Brightfield micrograph of a cross section of FGs and SGs stained with Oil Red O dye for the presence of fat granules. No positive signal is visible. Arrowheads show the ducts of the FGs. Inset: A higher magnification of the FG. Arrowheads show the duct of the FG. (B) The same section as in (A) shown under a fluorescence microscope. No fat granules were detected with Oil Red O staining. The cell nuclei (blue) were counterstained with DAPI. (C) A section through an anterior part of the larval abdomen stained with Oil Red O dye showing the fat granules in the perivisceral fat body lobes. (D) The same as in (C) under a fluorescent microscope. Fat granules stained with Oil Red O are clearly visible in the fat body lobes. Blue, cell nuclei stained with DAPI. (E) A transmission electron micrograph of a secretory cell from the FGs. No fat granules are found in the cytoplasm of the cell. (F) A transmission electron micrograph of an epithelial cell from the SG. No lipid granules are found in the cell cytoplasm. Scale bars: (A–D) = 25 µm; insets in (A,B) = 25 µm; (E,F) = 1 µm. Abbreviations: Cuticular intima (ci), fat body (fb), Filippi’s glands (Fg), midgut epithelium (gut), lumen of FGs and SG duct (lu), muscles of silk press (m), nucleus (n), silk glands (sg), secretory vesicles (sv).
4. Proteomic Analysis of Silk from FGs Ablated and Non-Ablated Larvae
To determine whether any specific proteins are added to the silk material from FGs, it performed a differential proteomic analysis of B. mori cocoons. It detected 2535 tryptic peptides within the reference in the UniProt database and identified more than 240 proteins (Supplementary Materials Table S2). Protein abundance was calculated using MaxQuant/Andromeda software (MPI of Biochemistry, Martinsried, Germany). Annotation of the identified proteins revealed that they included all previously known highly abundant structural silk proteins (fibroins, sericins, protease inhibitors or seroins) as well as with a number of less abundant enzymes and cellular proteins deposited in the silk by apocrine secretion. Comparison of the amounts of individual silk proteins from the cocoons of control and FGs ablated larvae revealed relatively minor differences (Supplementary Materials Table S2). Only two proteins showed statistically significant decreases in the cocoons of ablated larvae using Perseus analysis software, the enzyme apyrase (H9JTT3) and a small molecule of a Kazal-type trypsin protease inhibitor (P51902). While apyrase was 13-fold lower, the trypsin inhibitor appeared to be completely absent in the cocoons of larvae with ablated FGs. It was concluded that trypsin inhibitor may be involved in the regulation of proteolytic processing required for changes in adhesion strength or silk compactness.
5. Comparison of Proteomic Results with Previously Published Expression Data from a Sample of FG-Enriched Transcripts
Because the FGs are relatively small compared to the bulky SGs, candidate proteins from the FGs were expected to have rather low abundance in silk. To identify such candidate proteins with low abundance in the FGs, it compared proteomic results with previously published expression data from a sample of FG-enriched transcripts from wandering stage larvae [8]. It was found a total of 21 common proteins, as shown in the Venn diagram (Figure 7A). Only the proteins also included in the study by Wang et al. [9][8] were considered for further analysis (18 in total). Student’s t-test revealed no protein that was significantly decreased by ablation of FGs. However, the statistics were affected by missing values in the samples of treated larvae. This was true for ten proteins detected in no more than one cocoon of FG-ablated larvae. These ten proteins were added to the candidates (Table 1). Based on the SilkDB 3.0 platform [12][13], eight of the ten candidate proteins (Table 1) are predominantly expressed in SGs (including FGs); the remaining two proteins are less specific and mainly originate from the fat body. Not all candidate proteins also contained a signal peptide for secretion (Table 1). A total of 3 candidate proteins remained, including two enzymes and one protein with unknown function.
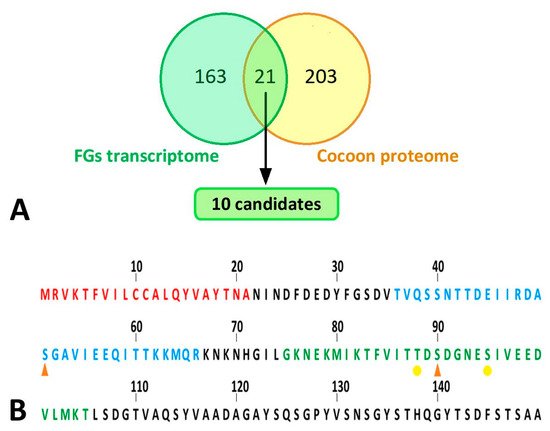
Figure 7. Bioinformatic analyses of silk proteins. (A) Comparison of results of proteomic analysis with published transcriptomic data. Yellow circle: all peptides detected by peptide mass fingerprinting; green circle: differentially expressed genes in the FG-enriched sample of wandering larvae spinnerets as published in Wang et al. [8]. The cut set contained a total of 21 proteins (18 of which were upregulated, 3 downregulated in FG-enriched transcriptome). This list of proteins was further narrowed down to 10 candidates (highlighted in green), which are listed in Table 1. (B) N-terminal part of B. mori FibH. Features indicated by letter colors: red—signal sequence; blue—peptide with significantly lower phosphorylation in larvae with ablated FGs; green—sequence not detected in FG-ablated larvae but containing phosphorylation in control larvae. Phosphorylation sites: orange arrowhead—major; yellow dot—minor.
Table 1. A definitive list of candidate proteins expressed in both the FG-enriched transcriptome [8] and proteins underrepresented in the silk of FG-depleted larvae. Data on their expression specificity were found in SilkDB 3.0 [12][13] and the presence of a signal peptide for secretion was predicted using SignalP 5.0 [14].
| Uniprot ID | SilkDB 3.0 ID | Expressed in SG/FG | Annotation | Signal Peptide |
|---|---|---|---|---|
| H9IWH6 | BGIBMGA001608 | Yes | Alpha-1,6-mannosyl-glycoprotein 2-beta-N-Acetylglucosaminyltransferase | No |
| H9J8H0 | BGIBMGA005812 | Yes | Arginine kinase | No |
| H9J9P6 | BGIBMGA006239 | No | Uncharacterized protein | Yes |
| H9JBW7 | BGIBMGA007012 | Yes | Extracellular serine/threonine protein kinase | No |
| H9JET3 | BGIBMGA008030 | Yes | Allantoate amidinohydrolase | No |
| H9JI83 | BGIBMGA009232 | No | Aldose 1-epimerase OS | No |
| H9JL76 | BGIBMGA010277 | Yes | Venom acid phosphatase Acph-1-like | Yes |
| H9JLC2 | BGIBMGA010323 | Yes | Uncharacterized protein OS | Yes |
| H9JPD2 | BGIBMGA011386 | Yes | Uncharacterized protein OS | No |
| H9JQ96 | BGIBMGA011702 | Yes | EN protein binding/engrailed nuclear Homeoprotein-regulated protein | No |
6. Identification and Quantification of Phosphorylation of Silk Proteins
Because two of the three secretory proteins described in the previous paragraph as candidates for differential presence of silks from FG ablated and control larvae were involved in the transfer or removal of phosphate groups, it compared the phosphorylation of the silk proteins using a mass spectrometric approach. The significance of differences was assessed in Perseus using the false discovery rate (Supplementary Materials Table S3, (q values)). Quantitative comparison of silk proteins between the control and ablated larval cocoons revealed a single site of significantly higher phosphorylation in the control group. This was a phospho-serine (S51-ph) within the sequence TVQSSNTTDEIIRDASGAVIEEQITTKKMQR originating from the region near the N-terminus of the FibH protein (Uniprot code P05790) (Figure 7B). Interestingly, another phospho-serine (S90-ph) GKNEKMIKTFVITTDSDGNESIVEEDVLMKT of FibH (Figure 7B) located near the first sequence also showed a similar trend. However, it could not be accurately quantified because no signal was detected in cocoons produced by ablated larvae. Overall, the N-terminus of FibH appeared to be phosphorylated at serine 51 in t cocoons of control larvae, and this phosphorylation was absent in cocoons from FG-ablated larvae.
References
- Sehadova, H.; Zavodska, R.; Zurovec, M.; Sauman, I. The Filippi’s Glands of Giant Silk Moths: To Be or Not to Be? Insects 2021, 12, 1040.
- Patra, S.; Singh, R.N.; Raziuddin, M. Morphology and histology of Lyonet’s gland of the tropical tasar silkworm, Antheraea mylitta. J. Insect Sci. 2012, 12, 1–7.
- Chen, F.; Porter, D.; Vollrath, F. Structure and physical properties of silkworm cocoons. J. R Soc. Interface 2012, 9, 2299–2308.
- Chen, F.J.; Porter, D.; Vollrath, F. Morphology and structure of silkworm cocoons. Mat. Sci. Eng. C Mater. 2012, 32, 772–778.
- Sehadova, H.; Guerra, P.A.; Sauman, I.; Reppert, S.M. A re-evaluation of silk measurement by the cecropia caterpillar (Hyalophora cecropia) during cocoon construction reveals use of a silk odometer that is temporally regulated. PLoS ONE 2020, 15, e0228453.
- Guerra, P.A.; Reppert, S.M. Dimorphic cocoons of the cecropia moth (Hyalophora cecropia): Morphological, behavioral, and biophysical differences. PLoS ONE 2017, 12, e0174023.
- Hamilton, C.A.; St Laurent, R.A.; Dexter, K.; Kitching, I.J.; Breinholt, J.W.; Zwick, A.; Timmermans, M.J.T.N.; Barber, J.R.; Kawahara, A.Y. Phylogenomics resolves major relationships and reveals significant diversification rate shifts in the evolution of silk moths and relatives. BMC Evol. Biol. 2019, 19, 182.
- Wang, X.; Li, Y.; Peng, L.; Chen, H.; Xia, Q.; Zhao, P. Comparative transcriptome analysis of Bombyx mori spinnerets and Filippi’s glands suggests their role in silk fiber formation. Insect Biochem. Mol. Biol. 2016, 68, 89–99.
- Wang, X.; Li, Y.; Liu, Q.; Tan, X.; Xie, X.; Xia, Q.; Zhao, P. GC/MS-based metabolomics analysis reveals active fatty acids biosynthesis in the Filippi’s gland of the silkworm, Bombyx mori, during silk spinning. Insect Biochem. Mol. Biol. 2019, 105, 1–9.
- Victoriano, E.; Pinheiro, D.O.; Gregorio, E.A. Histochemical and ultrastructural evidence of lipid secretion by the silk gland of the sugarcane borer Diatraea saccharalis (Fabricius) (Lepidoptera: Crambidae). Neotrop. Entomol. 2007, 36, 707–711.
- Engster, M.S. Studies on silk secretion in the trichoptera (F. Limnephilidae): I. Histology, histochemistry, and ultrastructure of the silk glands. J. Morphol. 1976, 150, 183–211.
- Wang, J.; Xia, Q.; He, X.; Dai, M.; Ruan, J.; Chen, J.; Yu, G.; Yuan, H.; Hu, Y.; Li, R.; et al. SilkDB: A knowledgebase for silkworm biology and genomics. Nucleic Acids Res. 2005, 33, D399–D402.
- Lu, F.; Wei, Z.Y.; Luo, Y.J.; Guo, H.L.; Zhang, G.Q.; Xia, Q.Y.; Wang, Y. SilkDB 3.0: Visualizing and exploring multiple levels of data for silkworm. Nucleic Acids Res. 2020, 48, D749–D755.
- Armenteros, J.J.A.; Tsirigos, K.D.; Sonderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.2K
Revisions:
2 times
(View History)
Update Date:
24 Dec 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




