Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Antonio Romero | + 2601 word(s) | 2601 | 2021-12-13 09:21:12 | | | |
| 2 | Rita Xu | Meta information modification | 2601 | 2021-12-23 03:03:38 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Romero, A. N-Tailed Monomeric Proto-Type-like Galectin. Encyclopedia. Available online: https://encyclopedia.pub/entry/17451 (accessed on 07 February 2026).
Romero A. N-Tailed Monomeric Proto-Type-like Galectin. Encyclopedia. Available at: https://encyclopedia.pub/entry/17451. Accessed February 07, 2026.
Romero, Antonio. "N-Tailed Monomeric Proto-Type-like Galectin" Encyclopedia, https://encyclopedia.pub/entry/17451 (accessed February 07, 2026).
Romero, A. (2021, December 22). N-Tailed Monomeric Proto-Type-like Galectin. In Encyclopedia. https://encyclopedia.pub/entry/17451
Romero, Antonio. "N-Tailed Monomeric Proto-Type-like Galectin." Encyclopedia. Web. 22 December, 2021.
Copy Citation
Galectins are multi-purpose effectors acting via interactions with distinct counterreceptors based on protein-glycan/protein recognition. These processes are emerging to involve several regions on the protein so that the availability of a detailed structural characterization of a full-length galectin is essential.
β-hairpin
β-sandwich
blood group B
1. Introduction
Storage of biological information involves more than nucleic acids and proteins. The ubiquity of occurrence, the enormous diversity already at the level of oligomers and the fine-tuned spatiotemporal regulation of the appearance of distinct structures are solid arguments for a fundamental functional meaning of the glycan part of cellular glycoconjugates [1][2][3][4][5][6]. Indeed, by molecular complementarity of oligosaccharides with a contact region in the carbohydrate recognition domains (CRDs) of sugar-binding proteins (lectins), glycan-encoded messages are ‘read’ and ‘translated’ into cellular effects [6][7][8]. Toward this end, triggering specific bioeffects, not only the selection of the binding partner(s), appears to matter. Furthermore, the lectin’s design, modularity and quaternary structure are first revealed in the case of the tetrameric leguminous lectin concanavalin A by a lower extent of crosslinking of certain cell surface receptors [9][10]. Fittingly, the context of presentation of the CRD shows a wide range of variability within lectin families. When considering the emerging multifunctionality of lectins, regions not involved in glycan binding can also affect their mode of action. Variability in design and the potential of regions beyond the glycan-binding site to be a physiologically relevant call for a detailed structural analysis within the lectin families in all their naturally occurring forms.
Focusing on the adhesion/growth-regulatory ga(lactose-binding) lectins, their common CRD is presented in three types of protein architecture, i.e., as (non)covalently associated homo/heterodimers (proto or tandem-repeat types) or as the chimera-type galectin-3 (Gal-3) with its N-terminal stalk attached to the CRD [11][12][13][14]. This highly dynamic, over 100-amino-acid-long sequence is composed of sections of known functionality; i.e., non-triple helical collagen-like repeats (for self-association) and an N-terminal peptide with two sites for serine phosphorylation (for intracellular compartmentalization) [12][15][16][17]. Two members of this lectin family are peculiar: galectin-related protein (GRP) and rat galectin-5 (rGal-5) present a short N-terminal extension (of up to 37 amino acids) of the canonical CRD of unknown function, which is clearly a challenge to study. Their special status as N-tailed proto-type-like proteins thus prompted to accomplish structural characterization of the full-length protein. In particular, it is of interest to define the structural features of the N-terminal extension, if adopted. Since respective attempts had so far been unsuccessful in the cases of human and chicken GRP, which had been crystallized as a truncated version [18][19], rGal-5 is the remaining target protein to try a full characterization of an N-tailed proto-type-like galectin.
This lectin was first purified from rat lung (denoted as RL-18) [20]. Sequencing of the cDNA from a rat reticulocyte library identified a strong homology (over 80%) to the C-terminal CRD of the tandem-repeat-type galectin-9 (Gal-9C), and monitoring among mammalian genomes disclosed its status as being uniquely present in rats [21][22][23][24]. The exon profile assumed that the rGal-5 gene originated from a species-specific gene duplication event followed by partial deletion to maintain the first exon coding for 13 amino acids and then the three exons of Gal-9C [12][24] (Figure 1). Notably, duplications and copy number variability of the galectin genes between species are not uncommon among mammals [25], rGal-5 being a specially processed species-specific form.
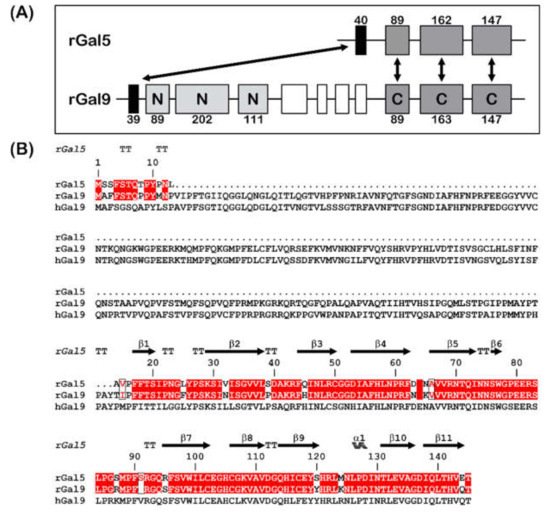
Figure 1. (A) Gene structures for rGal-5 and -9. The size of the exons (boxes) is indicated, and introns are drawn as lines (not drawn to scale). The N- and C-terminal CRDs of rGal-9 are labelled and given in different grey levels. Homologous exons are indicated (double arrows). (B) Sequence alignment of rGal-5 with rat and human Gal-9. Strictly conserved residues (red) and similar residues (boxed red letters) between rat proteins are shown. The upper lane represents the secondary structure elements of rGal-5 (α represents α-helices, β represents β-sheets and TT represents β-turns).
In solution, the current status of analysis describes rGal-5 as a monomer with a weak haemagglutinin activity [20][22][26]. Non-sialylated glycan termini are binding partners, especially when clustered, as is the case for N-acetyllactosamine (LacNAc) of the three complex-type N-glycans of the nonavalent pan-galectin-binding glycoprotein asialofetuin, and rGal-5 binding distinguishes late-stage apoptotic from secondary necrotic peripheral blood lymphocytes [27][28][29][30]. Selectivity in glycan binding is also implied in rGal-5′s involvement in the sorting processes during reticulocyte membrane remodeling by exosomal release [31].
2. Glycan Array Data
rGal-5 is first tested to determine its binding profile to chip-presented substances, mostly glycans up to the molecular mass of bacterial polysaccharides. By using an array platform with 609 compounds, the spacered histo-blood-group B (type 2) tetrasaccharide, LacNAc-based dimers and the xenoantigen with α1,3-linked galactose added to a LacNAc core were found to be frontrunners in terms of signal intensity, together with several bacterial polysaccharides (Figure 2). rGal-5, in contrast to GRP, which has lost the ability to bind β-galactosides [19], thus presents a profile with typical selectivity among this class of glycans. To report the contact pattern between rGal-5 and the selected carbohydrate ligands, we then carried out systematic screening to find conditions for crystallization. In these experimental series, we used the full-length protein to obtain structural information on the N-terminal tail.
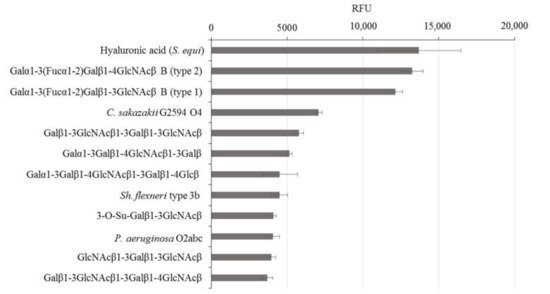
Figure 2. Top-12 glycans in the composition of the glycan array that exhibit binding with rGal-5.
3. Overall Crystallographic Structure of Full-Length rGal-5
Ligand-free full-length rGal-5 crystallizes in the monoclinic P21 space group and diffracts to a resolution of 1.7 Å. An estimation of the crystal solvent content suggested the presence of six galectin molecules in the asymmetric unit (Figure 3A). The rGal-5–lactose complex crystals belong to space group P22121, with only two molecules present in the asymmetric unit (Figure 4A), and diffract to a 1.9 Å resolution.
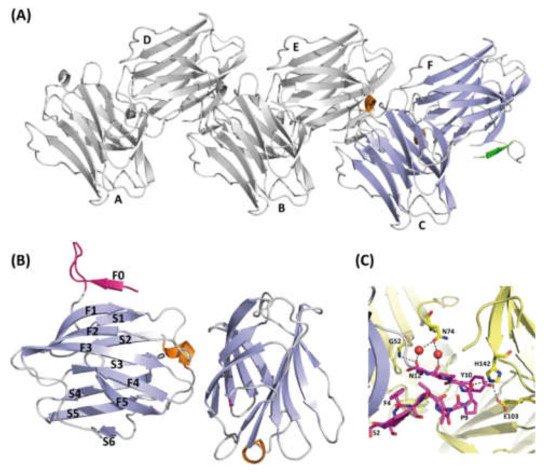
Figure 3. Structure of ligand-free rGal-5. (A) Ribbon diagram of the asymmetric unit of the crystals containing six molecules(A–F) of rGal-5 (helix in orange, β-strands in blue and the N-terminal extension in green). (B) Strands (labelled F0 to F5 on one side and S1 to S6 on the other) forming the characteristic β-sheets are labelled (helix in orange, β-strands in green and the N-terminal extension in magenta). An extra strand is found, named F0, placed immediately in front of a loop protruding from the CRD. (C) Intermolecular interactions stabilize the extended loop conformation. These interactions involve residues Gly52 (G52), Pro9 (P9), His142 (H142), Tyr10 (Y10), Glu103 (E103), Asn12 (N12), Gly52 (G52), Asn74 (N74). Symmetry-related molecules are shown in yellow.
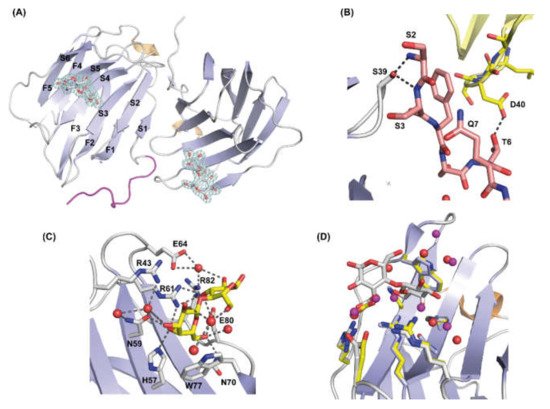
Figure 4. Structure of the rGal-5–lactose complex. (A) Overall architecture of the asymmetric unit of the rGal-5–lactose complex, with two CRDs; in one of them, the N-terminal residues (in magenta) adopt an extended geometry interacting with the edge of the β-sandwich. Strands that form the characteristic β-sheets are labelled (S1 to S6 on one strand, F1 to F5 on the other), and lactose molecules represented in sticks showing the 1.9 Å resolution 2Fo-Fc electron density map (in blue) contoured at 1.0 σ. (B) Inter- and intra-molecular interactions that stabilize the extended conformation of the N-terminal residues. These interactions involve residues Ser2 (S2), Ser3 (S3), Ser39 (S39), Thr6 (T6) and Asp40 (D40). Symmetry-related molecules are represented in different colors. (C) Close-up view of the ligand-binding site of rGal-5 to show interactions between the protein and lactose. Key protein residues [His57 (H57), Asn59 (N59), Arg61 (R61), Asn70 (N70), Glu80 (E80) Arg82 (R82), Arg43 (R43), Glu64 (E64) and Trp77 (W77)] and lactose are represented in stick mode and water molecules as red spheres. (D) Superposition of the ligand-binding sites of rGal-5 (yellow) and the rGal-5–lactose complex (grey). Side-chain positions of residues at this site are not affected by ligand binding, indicating a preformed geometry. In the ligand-free structure, the position of the residues is kept by interactions with water or glycerol molecules. Water molecules from this last structure are shown in purple for clarity.
The overall fold in ligand-free (Figure 3B) and -loaded rGal-5 (Figure 4A) is composed of two antiparallel β-sheets (F1 to F5 and S1 to S6 strands) that form the characteristic β-sandwich structure. A short 310 helix is placed between strands F5 and S2. Beyond analyzing the architecture of the contact site for glycans (see below), these crystals offered the opportunity to examine whether the N-terminal extension presents well-ordered elements or high flexibility.
4. Structure of Ligand-Free rGal-5
In the ligand-free structure, six rGal-5 molecules are arranged in the asymmetric unit, as shown in Figure 3A. The core of the different protein units can readily be superimposed onto each other, as revealed by the low average root mean square deviation (RMSD) value among them of 0.26 Å for all Cα atoms. Differences are attributed mainly to the N-terminus, and the six protein monomers can be divided into two groups (Figure 3A), based on the experimental electron density. In the first group (chains A–C), the 11 residues at the N-terminus are not visible in the electron density map. Their likely extended and flexible structure in solution can indeed be derived from our SAXS data: the ab initio model of rGal-5 calculated on this basis exhibits, expectably, a globular shape. Most interestingly, though, a cylindrical extension on its top was seen in the model. When placing the CRD within the spherical region, the extended N-terminal section matches the geometry of the cylindrical part of the SAXS model (see below).
When inspecting the second group (chains D–F), the electron density for this peptide stretch was clearly observed: the three amino acids from Ser2 to Ser5 form an extra β-strand, named F0, running in antiparallel direction to the C-terminal F1 strand. Residues Thr6 to Asn12 are in a loop, placed in parallel to the axis of the β-sandwich and protruding more than 10 Å from the S1 and F0 strands (Figure 3B).
The conformation within this loop is stabilized by interactions with symmetry-related molecules: hydrogen bonds between Pro9–His142, Tyr10–Glu103 and Asn12–Gly52 as well as a water bridge between Asn12 and Asn74 (Figure 3C).
5. Structure of Ligand-Loaded rGal-5
The two CRDs present in the asymmetric unit, which have bound lactose (Lac), exhibit very similar features to ligand-free rGal-5 (Figure 4A), with an RMSD value of only 0.3 Å for all Cα atoms. One of the two monomers in the asymmetric unit exhibits strong electron density for the first 10 residues so that their structure could be modelled. Intriguingly, these residues run parallel to the edge of the β-sandwich (Figure 4A) instead of forming the F0 strand and the protuberant loop observed in the ligand-free state. Intramolecular (hydrogen bonds between Ser2 and Ser3 with Ser39) and intermolecular contacts with symmetry-related molecules (hydrogen bond between Thr6 and Asp40) stabilize this special spatial arrangement (Figure 4B).
The carbohydrate-binding site in the concave face of the β-sheet is constituted by β-strands S4 to S6. The amino acids of the signature sequence, i.e., His57, Asn59, Arg61, Asn70, Glu80 and Arg82, directly interact with lactose through hydrogen bonding interactions. Additionally, Arg43, Gln45 and Glu64 form water-mediated hydrogen bonds with the ligand. As commonly found in galectin–lactose complexes, the indole ring of Trp77 stacks to the β-face of the pyranose ring of galactose (Figure 4C). In the absence of lactose, these residues form contacts with water (or glycerol molecules under conditions used for crystallization) molecules. Only minor rearrangements are observed for residues Arg43 and Glu64, which interact through water molecules with lactose (Figure 4D). Moving beyond defining the contact pattern, the thermodynamics of the ligand binding was analyzed by ITC.
6. ITC Measurements
rGal-5 interact with Lac with a dissociation constant of 136 ± 16 μM, which is lowered in the case of LacNAc to 30.5 ± 1.9 μM and 5.5 ± 0.6 μM for the blood-group B tetrasaccharide (Table 1). This stepwise affinity enhancement can be explained by the increased number of contacts that these ligands make with additional amino acids. In this case, we used its complex with an avian galectin cGRIFIN (see below) shown in Figure 5 and the respective model building to obtain the relevant information for the new additional contacts, as described below, between the tetrasaccharide and rGal5.
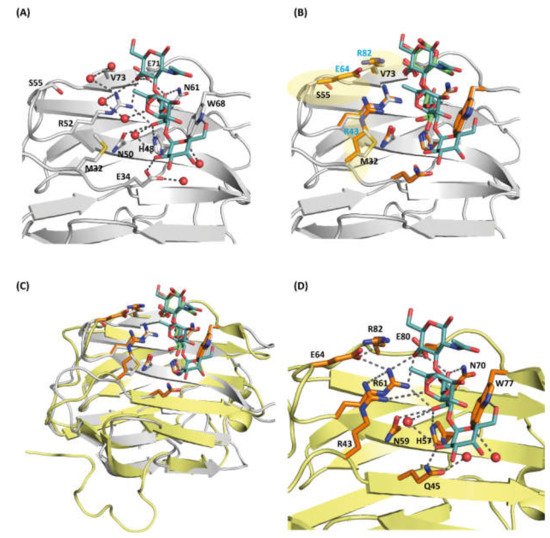
Figure 5. (A) Close-up view of the carbohydrate-binding site (CBD) of cGRIFIN showing the interactions between the histo-blood-group B tetrasaccharide and the active site residues [Met32 (M32), Glu34 (E34), His48 (H48), Asn50 (N50), Arg52 (R52), Ser55 (S55), Asn61 (N61), Trp68 (W68), Glu71 (E71) and Val73 (V73)]. (B) Structural comparison between cGRIFIN bound to the blood-group B tetrasaccharide complex (carbons in white) with the rGal-5/lactose complex (lactose in green, carbons of the interacting residues in orange). While residues such as His57, Asn59, Arg61, Asn70, Glu80 and Arg82 occupy almost the same position than residues in the former structure, the presence of charged residues (Arg53, Glu64 and Arg82) (highlighted in semitransparent yellow color) may lead to direct or water-mediated interactions with the ligand. (C) Superposition of the cGRIFIN/tetrasaccharide structure (grey; the histo-blood-group B tetrasaccharide in cyan) with the rGal-5–lactose complex (yellow; lactose in green) highlighting the differences in the active site residues between cGRIFIN and rGal-5 [Met32 (M32) to Arg43 (R43), Ser55 (S55) to Glu64 (E64), and Val73 (V73) to Arg82 (R82)]. (D) The rGal-5/histo-blood-group B tetrasaccharide modeled by superposition with the cGRIFIN/tetrasaccharide structure. The active site residues involved in bindig of the ligand are: Arg43 (R43), Gln45 (Q45), His57 (H57), Asn59 (N59), Arg61 (R61), Glu64 (E64), Asn70 (N70), Trp77 (W77), Glu80 (E80) and Arg82 (R82). Residues Arg43, Glu64 and Arg82 might explain the affinity of rGal-5 for the histo-blood-group B (type 2) tetrasaccharide.
Table 1. ITC data for ligand binding to recombinant rGal-5 (at 25 °C).
| Ligand | Kd (μM) | Stoichiometry | ΔG0obs (kcal/mol) | ΔH0obs (kcal/mol) | −TΔS0obs (kcal/mol) |
|---|---|---|---|---|---|
| Lactose | 121 ± 5 | 0.95 ± 0.05 | −5.35 | −4.99 ± 0.08 | −0.36 |
| 151 ± 5 | 0.99 ± 0.06 | −5.22 | −5.03 ± 0.41 | −0.18 | |
| LacNAc | 28.6 ± 2.0 | 0.92 ± 0.01 | −6.1 | −10.0 ± 0.2 | 3.87 |
| 32.3 ± 7.1 | 0.94 ± 0.20 | −6.0 | −10.5 ± 2.6 | 4.43 | |
| Tetrasacc haride |
6.1 ± 0.2 | 0.94 ± 0.01 | −7.11 | −5.60 ± 0.03 | −1.51 |
| 5.0 ± 0.2 | 0.98 ± 0.01 | −7.11 | −5.74 ± 0.03 | −1.37 |
7. Structure of Ligand-Loaded cGRIFIN
Our attempts to crystallize rGal-5 bound to the blood-group B tetrasaccharide were unsuccessful. Thus, we decided to test chicken GRIFIN (cGRIFIN) as a model for the binding of this compound. This very stable protein has been previously crystallized in several conditions [32]. We were able to obtain crystals of cGRIFIN in the presence of the blood-group B tetrasaccharide. These crystals diffracted up to a resolution of 1.14 Å. This high-resolution data allowed us to build the sugar structure in the electron density in both carbohydrate-binding sites of the dimer. A comparison of the lactose-bound (PDB 5NLE) and the tetrasaccharide-bound cGRIFIN structures shows the absence of any significant structural change between these two structures, the RMSD value being 0.382 Å for all Cα atoms. The GalB moiety fully superposes with the galactose moiety of lactose, forming H-bonds with His46, Asn48, Arg50, Asn59 and Glu69. On the other hand, the GlcNacB moiety is rotated in the tetrasaccharide compared to the glucose moiety of lactose. Despite this change in the conformation, the H-bond with Glu69 is conserved. The acetamido group is exposed to the solvent as it is the FucA moiety. The GalA moiety establishes two additional H-bonds, one of them linking the 6′-hydroxyl group with the NE atom of Trp66. The second one extends the binding site beyond the S4 strand, linking the 2′-hydroxyl group with Glu32 (Figure 5A).
The superposition of the lactose-bound rGal-5 structure with the blood-group B tetrasaccharide-bound cGRIFIN gave an RMSD of 0.69 Å for all Cα atoms, showing the similarity of both complexes. This similarity allows us to analyze the interactions that could be established between this ligand and rGal-5 (Figure 5B). The GalB and GlcNacB moieties of the tetrasaccharide could stablish the same interactions as those observed for lactose, including the one with Arg82. The GalA moiety is properly placed to interact with the NE atom of Trp66 and with the side chain of Gln45, a residue from the S3 strand. In addition, Arg43 faces the FucA ring and could interact with this moiety, expanding the ligand-protein surface of contact (Figure 5C). This last residue belongs to the loop connecting the S3 and F2 strands, the region interacting with the N-terminal residues in the ligand bound rGal-5 structure.
References
- Gabius, H.-J.; Roth., J. An introduction to the sugar code. Histochem. Cell Biol. 2017, 147, 111–117.
- Corfield, A.P. Eukaryotic protein glycosylation: A primer for histochemists and cell biologists. Histochem. Cell Biol. 2017, 147, 119–147.
- Kopitz, J. Lipid glycosylation: A primer for histochemists and cell biologists. Histochem. Cell Biol. 2017, 147, 175–198.
- Suzuki, N. Glycan diversity in the course of vertebrate evolution. Glycobiology 2018, 29, 625–644.
- Cummings, R.D. Stuck on sugars: How carbohydrates regulate cell adhesion, recognition, and signaling. Glycoconj. J. 2019, 36, 241–257.
- Kaltner, H.; Abad-Rodríguez, J.; Corfield, A.P.; Kopitz, J.; Gabius, H.-J. The sugar code: Letters and vocabulary, writers, editors and readers and biosignificance of functional glycan-lectin pairing. Biochem. J. 2019, 476, 2623–2655.
- Kilpatrick, D.C. Animal lectins: A historical introduction and overview. Biochim. Biophys. Acta 2002, 1572, 187–197.
- Manning, J.C.; Romero, A.; Habermann, F.A.; García Caballero, G.; Kaltner, H.; Gabius, H.-J. Lectins: A primer for histochemists and cell biologists. Histochem. Cell Biol. 2017, 147, 199–222.
- Gunther, G.R.; Wang, J.L.; Yahara, I.; Cunningham, B.A.; Edelman, G.M. Concanavalin A derivatives with altered biological activities. Proc. Natl. Acad. Sci. USA 1973, 70, 1012–1016.
- Ludwig, A.-K.; Kaltner, H.; Kopitz, J.; Gabius, H.-J. Lectinology 4.0: Altering modular (ga)lectin display for functional analysis and biomedical applications. Biochim. Biophys. Acta 2019, 1863, 935–940.
- Kasai, K.-I.; Hirabayashi, J. Galectins: A family of animal lectins that decipher glycocodes. J. Biochem. 1996, 119, 1–8.
- Cooper, D.N.W. Galectinomics: Finding themes in complexity. Biochim. Biophys. Acta 2002, 1572, 209–231.
- Kaltner, H.; Toegel, S.; García Caballero, G.; Manning, J.C.; Ledeen, R.W.; Gabius, H.-J. Galectins: Their network and roles in immunity/tumor growth control. Histochem. Cell Biol. 2017, 147, 239–256.
- García Caballero, G.; Kaltner, H.; Kutzner, T.J.; Ludwig, A.-K.; Manning, J.C.; Schmidt, S.; Sinowatz, F.; Gabius, H.-J. How galectins have become multifunctional proteins. Histol. Histopathol. 2020, 35, 509–539.
- Hughes, R.C. Mac-2: A versatile galactose-binding protein of mammalian tissues. Glycobiology 1994, 4, 5–12.
- Gao, X.; Liu, J.; Liu, X.; Li, L.; Zheng, J. Cleavage and phosphorylation: Important post-translational modifications of galectin-3. Cancer Metastasis Rev. 2017, 36, 367–374.
- Romero, A.; Gabius, H.-J. Galectin-3: Is this member of a large family of multifunctional lectins (already) a therapeutic target? Expert Opin. Ther. Targets 2019, 23, 819–828.
- Zhou, D.; Ge, H.; Sun, J.; Gao, Y.; Teng, M.; Niu, L. Crystal structure of the C-terminal conserved domain of human GRP, a galectin-related protein, reveals a function mode different from those of galectins. Proteins 2008, 71, 1582–1588.
- García Caballero, G.; Flores-Ibarra, A.; Michalak, M.; Khasbiullina, N.; Bovin, N.V.; André, S.; Manning, J.C.; Vértesy, S.; Ruiz, F.M.; Kaltner, H.; et al. Galectin-related protein: An integral member of the network of chicken galectins. 1. From strong sequence conservation of the gene confined to vertebrates to biochemical characteristics of the chicken protein and its crystal structure. Biochim. Biophys. Acta 2016, 1860, 2285–2297.
- Cerra, R.F.; Gitt, M.A.; Barondes, S.H. Three soluble rat β-galactoside-binding lectins. J. Biol. Chem. 1985, 260, 10474–10477.
- Jung, S.K.; Fujimoto, D. A novel β-galactoside-binding lectin in adult rat kidney. J. Biochem. 1994, 116, 547–553.
- Gitt, M.A.; Wiser, M.F.; Leffler, H.; Herrmann, J.; Xia, Y.; Massa, S.M.; Cooper, D.N.W.; Lusis, A.J.; Barondes, S.H. Sequence and mapping of galectin-5, a β-galactoside-binding lectin, found in rat erythrocytes. J. Biol. Chem. 1995, 270, 5032–5038.
- Wada, J.; Kanwar, Y.S. Identification and characterization of galectin-9, a novel β-galactoside-binding mammalian lectin. J. Biol. Chem. 1997, 272, 6078–6086.
- Lensch, M.; Lohr, M.; Russwurm, R.; Vidal, M.; Kaltner, H.; André, S.; Gabius, H.-J. Unique sequence and expression profiles of rat galectins-5 and -9 as a result of species-specific gene divergence. Int. J. Biochem. Cell Biol. 2006, 38, 1741–1758.
- Kaltner, H.; Raschta, A.-S.; Manning, J.C.; Gabius, H.-J. Copy-number variation of functional galectin genes: Studying animal galectin-7 (p53-induced gene 1 in man) and tandem-repeat-type galectins-4 and -9. Glycobiology 2013, 23, 1152–1163.
- André, S.; Kaltner, H.; Lensch, M.; Russwurm, R.; Siebert, H.-C.; Fallsehr, C.; Tajkhorshid, E.; Heck, A.J.R.; von Knebel-Döberitz, M.; Gabius, H.-J.; et al. Determination of structural and functional overlap/divergence of five proto-type galectins by analysis of the growth-regulatory interaction with ganglioside GM1 in silico and in vitro on human neuroblastoma cells. Int. J. Cancer 2005, 114, 46–57.
- Leffler, H.; Barondes, S.H. Specificity of binding of soluble rat lung lectins to substituted and unsubstituted mammalian β-galactosides. J. Biol. Chem. 1986, 261, 10119–10126.
- Dam, T.K.; Gabius, H.-J.; André, S.; Kaltner, H.; Lensch, M.; Brewer, C.F. Galectins bind to the multivalent glycoprotein asialofetuin with enhanced affinities and a gradient of decreasing binding constants. Biochemistry 2005, 44, 12564–12571.
- Wu, A.M.; Singh, T.; Wu, J.H.; Lensch, M.; André, S.; Gabius, H.-J. Interaction profile of galectin-5 with free saccharides and mammalian glycoproteins: Probing its fine-specificity and the effect of naturally clustered ligand presentation. Glycobiology 2006, 16, 524–537.
- Beer, A.; André, S.; Kaltner, H.; Lensch, M.; Franz, S.; Sarter, K.; Schulze, C.; Gaipl, U.S.; Kern, P.; Herrmann, M.; et al. Human galectins as sensors for apoptosis/necrosis-associated surface changes of granulocytes and lymphocytes. Cytom. A 2008, 73, 139–147.
- Barrès, C.; Blanc, L.; Bette-Bobillo, P.; André, S.; Mamoun, R.; Gabius, H.-J.; Vidal, M. Galectin-5 is bound onto the surface of rat reticulocyte exosomes and modulates vesicle uptake by macrophages. Blood 2010, 115, 696–705.
- Ruiz, F.M.; Gilles, U.; Ludwig, A.-K.; Sehad, C.; Shiao, T.C.; García Caballero, G.; Kaltner, H.; Lindner, I.; Roy, R.; Reusch, D.; et al. Chicken GRIFIN: Structural characterization in crystals and in solution. Biochimie 2018, 146, 127–138.
More
Information
Subjects:
Biophysics
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
690
Revisions:
2 times
(View History)
Update Date:
23 Dec 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




