Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Dacheng Liang | + 3517 word(s) | 3517 | 2021-12-03 02:59:01 | | | |
| 2 | Bruce Ren | Meta information modification | 3517 | 2021-12-22 02:01:51 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Liang, D. Root-to-Shoot Long-Distance Mobile miRNAs Identified from Nicotiana Rootstocks. Encyclopedia. Available online: https://encyclopedia.pub/entry/17395 (accessed on 14 January 2026).
Liang D. Root-to-Shoot Long-Distance Mobile miRNAs Identified from Nicotiana Rootstocks. Encyclopedia. Available at: https://encyclopedia.pub/entry/17395. Accessed January 14, 2026.
Liang, Dacheng. "Root-to-Shoot Long-Distance Mobile miRNAs Identified from Nicotiana Rootstocks" Encyclopedia, https://encyclopedia.pub/entry/17395 (accessed January 14, 2026).
Liang, D. (2021, December 21). Root-to-Shoot Long-Distance Mobile miRNAs Identified from Nicotiana Rootstocks. In Encyclopedia. https://encyclopedia.pub/entry/17395
Liang, Dacheng. "Root-to-Shoot Long-Distance Mobile miRNAs Identified from Nicotiana Rootstocks." Encyclopedia. Web. 21 December, 2021.
Copy Citation
Root-derived mobile signals play critical roles in coordinating a shoot’s response to underground conditions. However, the identification of root-to-shoot long-distance mobile signals has been scant.
long-distance transport
mobile miRNA
root-to-shoot
interfamilial graft
1. Introduction
One of the most fascinating aspects of plant grafting is the movement of macromolecules between a scion and rootstock (e.g., small RNA-mediated long-distance silencing movement [1][2], protein shuttling [3] and genetic material transfer [4][5][6][7][8]). The macromolecule movement may occur locally through cell-to-cell transfer via plasmodesmata (PD) or systematically through vasculature-mediated inter-tissue translocation. There are two specialized vascular tissues, namely the phloem and xylem, that usually serve as the superhighway for the long-distance transport of water, photoassimilates, nutrients, minerals and other signaling molecules. Emerging studies show that macromolecules are also moving through these superhighways. For instance, the florigen signal FLOWERING LOCUS T (FT) is generated in leaf tissue but is transmitted to the floral meristem via phloem [9][10][11][12]. This movement is critical for plants to exert their reproductive functions in response to environmental input, which in this case is light. However, new studies have found a certain type of transfer from a scion to rootstock may be physically inconclusive. For example, some proteins that are targeted toward suborganelles are also loaded into phloem and transmitted down to the rootstock, and the chloroplast-localized ferredoxin-NADPH oxidoreductase fused with GFP controlled by the 35S promoter in the scion was found down to the root meristem of a non-transgenic rootstock [13]. Further experiments have found those proteins during their transit to their target suborganelles, rather than via a secreted pathway, can be engulfed into the phloem stream and brought into the rootstock [13], implying that some proteins could be passively transported via phloem during the process of transit.
The passive long-distance transport could also occur to RNA. The majority of the RNAs with abundant expression and longer half-lives were detected as mobile RNAs, and only a small proportion were identified as the low-abundance mobile RNAs, suggesting the passive movement mechanism plays an important role in the intraspecific scion–rootstock exchange [14]. Abundance-driven RNA mobility was also observed in the parasite–host system (e.g., the Arabidopsis–Cuscuta and tomato–Cuscuta interaction) [15]. RNAs in high abundance from either the host or parasite tend to be detected in the tissues from the respective opponent, implying that the movement of highly abundant RNAs might not be selective.
Aside from the abundance model, the structural model provides another mechanism for intercellular RNA movement. A tRNA-like structure is capable of moving between cells, such that the non-mobile GUS mRNA can be, when adorned at the 5′ UTR, transmitted to the shoot tip in the grafting assay [16]. These insightful results could spur further investigation on how this particular structure can impart mRNA movement and its underlying mechanisms.
Apparently, the small RNAs, typically 20–24 nt and including small interfering RNA (siRNA) and miRNA [17], do not possess the tRNA-like structure to perform non-autonomous movement. For example, siRNA-mediated post-transcript gene silencing can move from root to shoot or shoot to root without the help of a special RNA structure [1][2][18]. However, the genetic components for small RNA processing, particularly RNA amplification, are required in the recipient tissues (e.g., DICER-like 3 (DCL3) or RNA-dependent RNA polymerase (RDR2)) [1] and also in the sending tissues (e.g., RDR6) [2]. These results further emphasized that some small RNAs could make their move in a quantity-dependent manner.
On the contrary, the endogenous miRNAs are RDR-independent small RNAs (sRNAs) and tend to exert their functions locally [19][20]. However, many studies have shown a good number of miRNAs can perform long-distance movement from a scion to a rootstock [21][22]. We need to point out that so far, the systemically mobile miRNAs have been validated through overexpression of the target miRNA in the scion or examination in a rootstock hen1 mutant background that is unable to accumulate mature miRNAs. These verifying methods again alluded to the importance of abundance through artificial enrichment of the target miRNAs, thus compromising the biological significance of scion-to-rootstock mobile miRNAs. Questions arise as to whether any miRNA can perform long-distance movement without amplification or phloem streaming or if any miRNA can move from the rootstock to the scion, which is against the phloem stream.
2. Hypocotyl Grafting and Small RNA Deep Sequencing in At Scions and Nb Rootstocks
To stringently characterize the small RNAs that move from root to shoot, we adopted hypocotyl grafting, in which the graft union occurred at the hypocotyl region of At and Nb (Figure 1A). This type of graft, unlike those exploring the source-to-sink movement, separated the shoot and root spatially, thus avoiding the leaf source-derived signals being included. We found that the life span was extensively extended in the At/Nb grafts [23], and we hypothesized that any small RNAs from the rootstock could partially represent contributing factors to the observed phenotypic changes. With this premise, we collected the samples at the mature stage of the At/Nb graft or 90 days after grafting (DAG) for small RNA library construction. Because sample collection in a similar timescale is not applicable for At/At or Nb/Nb self-grafts, we chose the similarly developmental stage of self-grafts (e.g., 40–45 DAG) for a negative control.

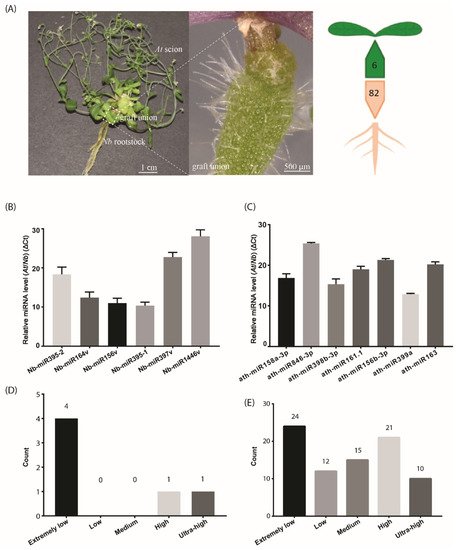
Figure 1. The number of identified mobile miRNAs in the Arabidopsis/Nicotiana interfamilial graft. (A) A representative At/Nb interfamilial graft showing the scion, rootstock and graft union. In total, 6 and 82 miRNAs were identified to move from root to shoot and shoot to root, respectively, in Nicotiana benthamiana and Arabidopsis thaliana. (B) Expression levels of the selected Nb miRNAs were determined in the At scion by quantitative real-time PCR. (C) Expression levels of the selected At miRNAs were determined in the Nb rootstock by quantitative real-time PCR. (D) Classification of expression level for the identified mobile miRNA in the scion. (E) Classification of expression level for the identified mobile miRNA in the rootstock. Data in (B,C) (mean ± standard deviation) were generated from three biological replicates and two technical replicates for each. U6 RNA was used as the internal reference. ΔCt values were the difference between the Ct values of the selected miRNA and U6 and thus were inversely proportional to the amount of the target miRNA in the samples.
Four groups of materials that were labeled as AGS (Arabidopsis grafting scion in At/Nb heterograft), ACS (Arabidopsis control scion in At/At homograft), NGR (Nicotiana grafting rootstock in At/Nb heterograft) and NCR (Nicotiana control rootstock in Nb/Nb homograft) were harvested for RNA deep sequencing. More than 13.85 million clean reads in the size of 18–30 nt for each sample were obtained by removing the low-quality reads (e.g., reads of a length <18 nt or >30 nt, reads with more than three unknown bases or reads without adaptors). Bowtie analysis was performed to determine the read distribution in the clean reads. The results showed that the unannotated reads accounted for more than 50% of all reads in all samples, and the miRNA reads accounted for about 6.3%, 4.4%, 1.9% and 2% in the AGS, ACS, NGR and NCR, respectively. More than 60% of the total sRNA reads including miRNA and unannotated RNAs in the AGS and ACS were further mapped to the Arabidopsis genome, and less than 1% of the remaining unmapped reads were re-mapped onto the Nb genome (Table 1). Similarly, in the Nb rootstock, about 0.29% and 0.05% of the total sRNA reads from NGR and NCR, respectively, were mapped to the Arabidopsis genome (Table 2). These results suggested that a small fraction of the total small RNAs could potentially move across the graft union.
Table 1. Deep sequencing of small RNA libraries from the At scion and mapping to the At and Nb genomes.
| Samples | Species | Tissue Samples | Total Reads | Clean Reads (%) | Total sRNA Reads from At | ||||
|---|---|---|---|---|---|---|---|---|---|
| Scion | Rootstock | sRNA Reads | Mapped sRNA Reads (%) | Unmapped sRNA Reads | Re-Mapping to Nb (%) | ||||
| AGS | A.thaliana | N.benthamiana | cauline leaf, stem, flower |
77,896,720 | 62,481,229 (80.21%) | 41,991,400 | 25,206,466 (60.03%) | 16,784,934 | 298,739 (0.71%) |
| ACS | A.thaliana | A.thaliana | cauline leaf, stem, flower |
76,187,700 | 57,001,571 (74.82%) | 34,161,421 | 20,766,148 (60.79%) | 13,395,273 | 237,220 (0.69%) |
Table 2. Deep sequencing of small RNA libraries from Nb rootstock and mapping to At and Nb genome.
| Sample | Species | Tissue Samples | Total Reads | Clean Reads (%) | Total sRNA Reads from Nb | ||||
|---|---|---|---|---|---|---|---|---|---|
| Scion | Rootstock | sRNA Reads | Mapped sRNA Reads (%) | Unmapped sRNA Reads | Re-Mapping to At (%) | ||||
| NGR | A.thaliana | N.benthamiana | root | 68,325,049 | 62,307,593 (91.19%) | 52,616,678 | 36,897,829 (70.13%) | 15,718,849 | 152,412 (0.29%) |
| NCR | N.benthamiana | N.benthamiana | root | 256,050,792 | 223,052,992 (87.11%) | 166,724,440 | 112,256,797 (67.33%) | 54,467,643 | 89,082 (0.05%) |
3. Small RNA Movement across the Graft Union
A very simple method to recover the high-confidence mobile sRNAs dictates that RNA reads from either the scion or rootstock that are mapped to the opposite part of the grafts should not appear in the corresponding part of self-grafts. Under this stringent purview, we recovered six candidates from Nb in the At scions (Table 3). These miRNAs were highly conserved, belonging to five miRNA families, including miR156, miR164, miR395, miR397 and miR1446. Since these miRNAs showed sequence variation to the existing miRNA members, they were referred to as Nb-miR156v, Nb-miR164v, Nb-miR395-1, Nb-miR395-2, Nb-miR397v and Nb-miR1446v, respectively, in this study (Table 3). The precursor sequences were also identified from the Nb genome. They may be ascribed as high-confidence root-to-shoot mobile candidates, as they repeatedly appeared in all heterografting samples.
Table 3. Mobile Nb miRNAs identified from the At scion in an At/Nb heterograft.
| Sequencing ID | miRNA Family | Mature miRNA Sequence | Length | AGS Read Counts | ACS Read Counts | ||||
|---|---|---|---|---|---|---|---|---|---|
| AGS1 | AGS2 | AGS3 | ACS1 | ACS2 | ACS3 | ||||
| conservative_Niben101Scf00647_2272 (Nb-miR156 variant or Nb-miR156v) | miR156 | UGACAGAAGAGAGUGGGC | 18 | 4 | 9 | 6 | 0 | 0 | 0 |
| conservative_Niben101Scf00747_2488 (Nb-miR164 variant or Nb-miR164v) | miR164 | UGGAGAAGCAGGGCACAUGC | 20 | 1 | 1 | 1 | 0 | 0 | 0 |
| conservative_Niben101Scf02279_7619 (Nb-miR395-1) | miR395 | CUGAAGUGUUUGGGGGAACUCU | 22 | 3 | 13 | 1 | 0 | 0 | 0 |
| conservative_Niben101Scf02027_6631 (Nb-miR1446 variant or Nb-miR1446v) | miR1446 | UUCUGAACUCUCUCCCUCAAU | 21 | 0 | 3 | 0 | 0 | 0 | 0 |
| conservative_Niben101Scf02778_9073 (Nb-miR397 variant or Nb-miR397v) | miR397 | UCAUUGAGUGCAGCGUUGAUGA | 22 | 1 | 3 | 5 | 0 | 0 | 0 |
| conservative_Niben101Scf01112_4153 (Nb-miR395-2) | miR395 | CUGAAGUGUUUGGGGGAACUCCG | 23 | 25 | 47 | 45 | 0 | 0 | 0 |
Likewise, the same selection criteria applied to the Nb rootstock led to the identification of 82 At miRNAs, a much higher number than those in the rootstock-to-scion migrating direction. This in turn agreed with the bulk flow in the source-to-sink direction. These potentially mobile miRNAs accounted for nearly one quarter of the total Arabidopsis miRNAs (miRbase), suggesting the movement of substantial miRNAs via the phloem. We further compared the miRNAs in the AGS samples with those in the Nb samples and found 31 At miRNAs that shared the exact sequence with those in the Nb. Thus, we could not determine their mobility in this study. This might further imply that the number of mobile miRNAs through phloem could potentially increase with an improved technique.
We also noticed that the two subsets of mobile miRNAs shared no essential overlaps. Although the miR156 family member appeared in both subsets (Table 3), the Nb miR156 sequence corresponded to the sense strand, while the At miR156 sequence recovered in the rootstock was located in the antisense strand, agreeing with previous studies showing that the miR156* strand could be detected in the phloem [24]. Taken together, the non-overlaps between the two mobile miRNA subsets implied the divergence of the regulatory mechanisms of the top-down and down-top miRNA movement.
4. Mobile miRNA Detection
To detect the mobile miRNAs, we adopted the stem-loop RT-qPCR procedure which proved to be specific and sensitive to miRNA detection [25]. In the scion tissues, five out of the six selected miRNAs were detected (Figure 1B), including Nb-miR395-1, -2, Nb-miR164v, Nb-miR156v, Nb-miR397v and Nb-miR1446v. In the rootstock, the seven selected miRNAs from At were detected (Figure 1C). All these detected miRNAs showed a very low accumulation relative to the U6 reference. We further checked the expression of these mobile miRNAs in the non-grafting tissues and found nearly one third of the root-to-shoot miRNAs (29%) belonged to the extremely low expression group (Figure 1D). In the shoot-to-root miRNA subset, nearly one third of them belonged to the extremely low expression group, and around 38% of the miRNAs were from the high or ultra-high expression group (Figure 1E), suggesting the abundance model [14] could partially explain the disparity in the miRNA numbers between the two subsets, given that the high expression miRNAs could easily be brought down to the rootstock through the phloem bulk flow.
5. Pre- and Mature miRNA, but Not the Pri-miRNA, Can Be Detected in the Scion or Rootstock
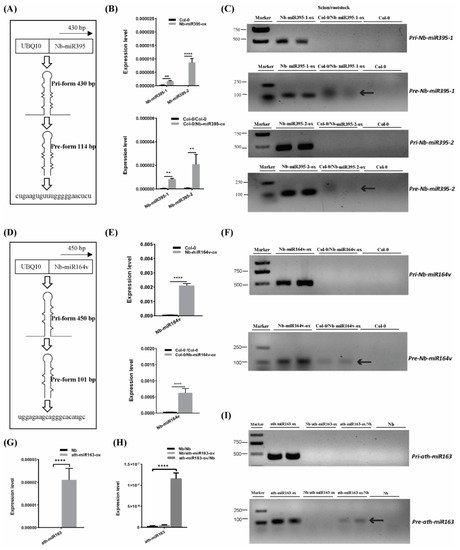
To further confirm the mobility of these miRNAs, we overexpressed the pri-miRNA coding sequence of Nb-miR395-1, Nb-miR395-2 and Nb-miR164v in Arabidopsis to check whether they sustained their mobility in Arabidopsis (Figure 2A,D). The stem-loop QPCR results revealed that the mature miRNAs were expressed in Arabidopsis and could be detected in the scion tissues in which the miRNA-overexpressing Arabidopsis was used as the rootstock (Figure 2B,E), suggesting the mature Nb-miR395-1, Nb-miR395-2 and Nb-miR164v could move to the scion as shown in the heterografts (Figure 1B). We found that the transcribed pri-miRNAs in all these selected miRNA loci (Nb-miR395-1, Nb-miR395-2 and Nb-miR164v) were beyond detection in the scions (Figure 2C,F), but the pre-miRNAs could be detected in the Arabidopsis scions (two independent materials), suggesting the pre-miRNAs together with the mature miRNAs could be transferred from the roots to the shoots. To further evaluate the specificity of the miRNA movement, we overexpressed an Arabidopsis-specific miRNA gene, miR163, which showed a potential for rootward mobility (Table S3) in N. benthamiana and used this transgenic Nb line (ath-miR163-ox) as the scion or rootstock (Figure 2G–I). QPCR showed that miR163 could not be detected in the wild-type (WT) Nb background but was highly expressed in the Nb transgenic line (Figure 2G). The grafting experiment revealed that miR163 from the Nb overexpressing line could be detected in the WT Nb rootstock rather than the Nb scion (Figure 2H), suggesting the specificity of its directional movement. Likewise, the pre-miRNA (but not the pri-miRNA) of miR163 could be detected in the WT Nb rootstock (Figure 2I), supporting the mobility of the miR163 precursor.

Figure 2. Pre- and mature miRNA detection in the scion. (A) Pri and pre forms of Nb-miR395-1. (B) Nb-miR395-1, -2 detection in the Arabidopsis overexpressing line (top) and in the WT Col-0 scion (bottom). (C) RT-PCR assays on pri and pre forms of Nb-miR395-1, -2. (D) Pri and pre forms of Nb-miR164v. (E) Nb-miR164v detection in the Arabidopsis overexpressing line (top) and in the WT Col-0 scion (bottom). (F) RT-PCR assays on pri and pre forms of Nb-miR164v. (G) QPCR quantification of ath-miR163 in the Nb overexpressing line. (H) QPCR quantification of ath-miR163 in the WT Nb scion and rootstock. (I) RT-PCR assays on pri and pre forms of ath-miR163 in the WT Nb scion and rootstock. The arrow indicates the precursor transcript in the WT scion or rootstock. The bars represent the means and standard deviations of six replicates (three biological replicates, each with two technical replicates). The two asterisks indicate p < 0.01, and four asterisks indicate p < 0.0001 (t-test).
6. Validation of miRNA Movement Using the MS2 System
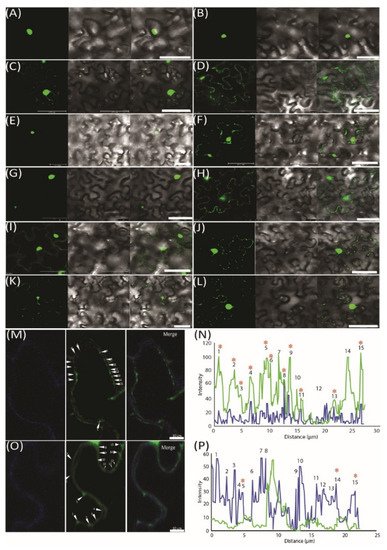
To further demonstrate the movement of miRNAs, we used an MS2 RNA visualizing system, which was adopted to visualize the RNA movement in the plants [26]. MS2FD-GFP was only detected in the nucleus when transiently co-expressed with the nonmobile RNA Actin2-SL24 [27] or SL24 blank vector (Figure 3A–C). To test how the root-to-shoot miRNAs could be mobile between cells, we fused two different forms of miRNAs (i.e., the pri and pre forms) with SL24. As seen in Figure 3E–G, the full-length pri forms of Nb-miR395-1 and Nb-miR397v were mainly accumulated in the nucleus, giving out a similar pattern to that of MS2FD-GFP or that of nonmobile RNAs (Figure 3A–C,E,G). However, the pre forms of Nb-miR395-1 and Nb-miR397v were localized to the punctate foci around the cell periphery (Figure 3F,H) in a pattern similar to that of FLOWERING LOCUS T (FT) mRNA FT-SL24 (Figure 3D). In pri-miR164-SL24, the GFP signal was simply uniformly detected at the cell periphery (Figure 3I). However, in pre-Nb-miR164v-SL24, the strong signal of the punctate green fluorescent foci appeared around the cell periphery (Figure 3J). Similarly, the mature Nb-miR164v and three tandem repeats of Nb-miR164v (3xNb-miR164v-SL24) could also be located in the punctate foci (Figure 3K,L), agreeing with the miRNA detection results (Figure 2E,F). To further confirm the punctate foci were overlapping with PD, we examined the localization of 3xNb-miR164V-SL24 GFP fluorescence and the signal of aniline blue that was used to indicate callose deposition at the PD neck, and we found the two signals were co-localized in the majority of the examined foci (Figure 3M,N). In contrast, the Actin2-SL24 GFP signal, shown in Figure 3O,P, was very scarce and only partly overlapping with the aniline blue signal (3 of 15 stains). These findings suggest that the mature and pre forms of the selected miRNAs were mobile and most probably targeted to PD to be transferred to the adjacent cell.

Figure 3. Pre form (rather than pri form) of root-to-shoot mobile miRNAs that were targeted toward plasmodesmata (PD). (A) MS2FD-GFP localization in the nucleus of Nb epidermal cells 2 days after Agrobacterium infiltration. (B) Co-infiltration of MS2FD-GFP and an SL24 empty vector. (C) Co-infiltration of MS2FD-GFP and an Actin2-SL24 negative control. (D) Co-infiltration of MS2FD-GFP and SL24-FT (FLOWERING LOCUS T) used as a positive control. (E) Pri-Nb-miR395-1 co-expressed with MS2FD-GFP. (F) Pre-Nb-miR395-1 co-expressed with MS2FD-GFP. (G) Pri-Nb-miR397v co-expressed with MS2FD-GFP. (H) Pre-Nb-miR397v co-expressed with MS2FD-GFP. (I) Pri-Nb-miR164v co-expressed with MS2FD-GFP. (J) Pre-Nb-miR164v co-expressed with MS2FD-GFP. (K) Mature Nb-miR164v fused with SL24 and its co-expression with MS2FD-GFP. (L) Three tandem repeats of Nb-miR164v (3xNb-miR164v-SL24) fused with SL24 and its co-expression with MS2FD-GFP. (M) Co-expression of 3xNb-miR164v-SL24 and MS2FD-GFP in aniline blue-stained N. benthamiana leaves. Left: image taken in the aniline blue channel. Middle: image taken in the GFP channel. Right: merged image from left and middle image. (N) Co-localization analysis of the GFP foci in 3xNb-miR164v-SL24 transiently expressing leaves. (O) Co-expression of non-mobile Actin2-SL24 RNA and MS2FD-GFP in aniline blue-stained N. benthamiana leaves. Left: image taken in the aniline blue channel. Middle: image taken in the GFP channel. Right: merged image from left and middle image. (P) Co-localization analysis of the GFP foci in Actin2-SL24 transiently expressing leaves. The numbers in (M–P) represent individual GFP spots. Co-localization of the GFP signal with aniline blue is indicated by a star sign. Scale bar in (A–L) represents 50 µm and scale bar in (M,O) represents 10 µm.
7. Phenotypic Modification in the Scion by Root-to-Shoot Mobile Nb-miR164v
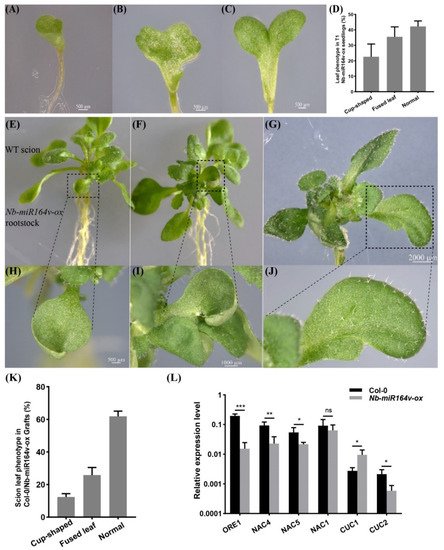
We next asked whether these root-to-shoot mobile miRNAs could cause any biological consequences on the scion morphology after moving into the scion. For this, we overexpressed Nb-miR395-1, Nb-miR395-2, Nb-miR397v, Nb-miR1446v and Nb-miR164v in Arabidopsis to check if they could cause any phenotypic changes. In the T2 generation of the Nb-miR395-1, Nb-miR395-2, Nb-miR397v and Nb-miR1446v overexpressing lines, no visual phenotype or developmental defects were observed. The grafting experiment involving these lines as rootstocks did not show any observable phenotypic difference with the Col-0 self-grafts. As a representative example, Nb-miR164v was chosen for further investigation, since the overexpression of Nb-miR164v in Arabidopsis gave rise to copious morphological defects, such as the fused and misshapen leaves (Figure 4A–D). These phenotypes were exactly similar to the Arabidopsis‘s own miR164 overexpressing lines [28][29][30], suggesting the similar role of Nb-miR164v in leaf development.

Figure 4. Phenotypic changes in a scion by root-to-shoot transmissible Nb-miR164v. (A–D) Overexpression of Nb-miR164v driven by a ubiquitin10 promoter in Arabidopsis, resulting in leaf defects such as the cup-shaped cotyledon (A), fully fused cotyledon (B) and partially fused cotyledon (C). (D) The percentage of Nb-miR-164v-ox seedlings showing the leaf phenotype as in (A–C). The bars represent the means and standard deviations of the two experiments (N = 50 in each experiment). (E–G) The WT Arabidopsis was used as a scion and grafted to the above plants with leaf defects. (H–J) Cup-shaped and partially fused leaves from the scion. Note that not all the leaves from the scion showed defects. (K) The percentage of grafts showing the altered leaf phenotype. The bars represent the means and standard deviations of three experiments (N = 15 in each experiment). (L) Expression of Nb-miR164v targets in the scion leaves. The bars represent the means and standard deviations of six replicates (three biological replicates, each with two technical replicates). *, **, and *** indicate p < 0.05, <0.001, and <0.0001, respectively.
Researchers then grafted the Nb-miR164v overexpressing line as a rootstock with the WT Col-0 scion. The first emerging leaves in these grafts showed the typical defects (Figure 4E–J), and the fusion of two different leaves could also occur (Figure 4G,J). In total, around 38% of the grafts showed the typical leaf phenotypes (Figure 4K). Since these defects are characteristic for plants with reduced CUC1 or CUC2 activity [28][29][30], we quantified the levels of the miR164 targets in the WT and the grafts. The results showed that the majority of the miR164 targets were significantly downregulated in the WT scion (Figure 4L), which was consistent with the expectations. These results strongly indicated that the Nb-miR164v in the rootstock moved into the scion to modify the leaf shape.
References
- Brosnan, C.A.; Mitter, N.; Christie, M.; Smith, N.A.; Waterhouse, P.M.; Carroll, B.J. Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 14741–14746.
- Liang, D.; White, R.; Waterhouse, P. Gene silencing in Arabidopsis spreads from the root to the shoot, through a gating barrier, by template-dependent, nonvascular, cell-to-cell movement. Plant Physiol. 2012, 159, 984–1000.
- Gallagher, K.L.; Sozzani, R.; Lee, C.M. Intercellular protein movement: Deciphering the language of development. Annu. Rev. Cell Dev. Biol. 2014, 30, 207–233.
- Lough, T.; Lucas, W. Integrative plant biology role of phloem long- distance macromolecular trafficking. Annu. Rev. Plant Biol. 2006, 57, 203–232.
- Stegemann, S.; Bock, R. Exchange of genetic material between cells in plant tissue grafts. Science 2009, 324, 649–651.
- Atkins, C.A.; Smith, P.M.; Rodriguez-Medina, C. Macromolecules in phloem exudates—A review. Protoplasma 2011, 248, 165–172.
- Fuentes, I.; Stegemann, S.; Golczyk, H.; Karcher, D.; Bock, R. Horizontal genome transfer as an asexual path to the formation of new species. Nature 2014, 511, 232.
- Yu, Y.; Jia, T.; Chen, X. The ‘how’ and ‘where’ of plant microRNAs. New Phytol. 2017, 216, 1002–1017.
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT Protein Movement Contributes to Long-Distance Signaling in Floral Induction of Arabidopsis. Science 2007, 316, 1030–1033.
- Lin, M.K.; Belanger, H.; Lee, Y.J.; Varkonyi-Gasic, E.; Taoka, K.; Miura, E.; Xoconostle-Cazares, B.; Gendler, K.; Jorgensen, R.A.; Phinney, B.; et al. FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cu-curbits. Plant Cell 2007, 19, 1488–1506.
- Yoo, S.; Hong, S.; Jung, H.; Ahn, J. The cotyledons produce sufficient FT protein to induce flowering: Evidence from cotyledon micrografting in Arabidopsis. Plant Cell Physiol. 2013, 54, 119–128.
- Zhu, Y.; Liu, L.; Shen, L.; Yu, H. NaKR1 regulates long-distance movement of FLOWERING LOCUS T in Arabidopsis. Nat. Plants 2016, 2, 16075.
- Paultre, D.; Gustin, M.; Molnar, A.; Oparka, K. Lost in transit: Long-distance trafficking and phloem unloading of protein signals in Arabidopsis homografts. Plant Cell 2016, 28, 2016–2025.
- Calderwood, A.; Kopriva, S.; Morris, R.J. Transcript Abundance Explains mRNA Mobility Data in Arabidopsis thaliana. Plant Cell 2016, 28, 610–615.
- Kim, G.; LeBlanc, M.L.; Wafula, E.K.; dePamphilis, C.W.; Westwood, J.H. Genomic-scale exchange of mRNA between a parasitic plant and its hosts. Science 2014, 345, 808–811.
- Zhang, W.; Thieme, C.J.; Kollwig, G.; Apelt, F.; Yang, L.; Winter, N.; Andresen, N.; Walther, D.; Kragler, F. tRNA-Related Sequences Trigger Systemic mRNA Transport in Plants. Plant Cell 2016, 28, 1237–1249.
- Axtell, M. Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 2013, 64, 137–159.
- Molnar, A.; Melnyk, C.W.; Bassett, A.; Hardcastle, T.J.; Dunn, R.; Baulcombe, D.C. Small Silencing RNAs in Plants Are Mobile and Direct Epigenetic Modification in Recipient Cells. Science 2010, 328, 872–875.
- Alvarez, J.P.; Pekker, I.; Goldshmidt, A.; Blum, E.; Amsellem, Z.; Eshed, Y. Endogenous and Synthetic MicroRNAs Stimulate Simultaneous, Efficient, and Localized Regulation of Multiple Targets in Diverse Species. Plant Cell 2006, 18, 1134–1151.
- Polydore, S.; Axtell, M.J. Analysis of RDR1/RDR2/RDR6-independent small RNAs in Arabidopsis thaliana improves MIRNA annotations and reveals unexplained types of short interfering RNA loci. Plant J. 2018, 4, 1051–1063.
- Marin-Gonzalez, E.; Suarez-Lopez, P. “And yet it moves”: Cell-to-cell and long-distance signaling by plant microRNAs. Plant Sci. 2012, 196, 18–30.
- Liu, L.; Chen, X. Intercellular and systemic trafficking of RNAs in plants. Nat. Plants 2018, 4, 869–878.
- Deng, Z.; Wu, H.; Jin, T.; Cai, T.; Jiang, M.; Wang, M.; Liang, D. A Sequential Three-Phase Pathway Constitutes Tracheary Element Connection in the Arabidopsis/Nicotiana Interfamilial Grafts. Front. Plant Sci. 2021, 12, 664342.
- Bhogale, S.; Mahajan, A.; Natarajan, B.; Rajabhoj, M.; Thulasiram, H.; Banerjee, A. MicroRNA156: A potential graft-transmissible microRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant Physiol. 2014, 164, 1011–1027.
- Varkonyi-Gasic, E.; Wu, R.; Wood, M.; Walton, E.F.; Hellens, R.P. Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 2007, 3, 12.
- Luo, K.-R.; Huang, N.-C.; Yu, T.-S. Selective Targeting of Mobile mRNAs to Plasmodesmata for Cell-to-Cell Movement. Plant Physiol. 2018, 177, 604–614.
- Thieme, C.; Rojas-Triana, M.; Stecyk, E.; Schudoma, C.; Zhang, W.; Yang, L.; Miñambres, M.; Walther, D.; Schulze, W.; Paz-Ares, J.; et al. Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants 2015, 1, 15025.
- Laufs, P.; Peaucelle, A.; Morin, H.; Traas, J. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 2004, 131, 4311–4322.
- Raman, S.; Greb, T.; Peaucelle, A.; Blein, T.; Laufs, P.; Theres, K. Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 2008, 55, 65–76.
- Berger, Y.; Harpaz-Saad, S.A.; Melnik, H.N.; Alvarez, J.P.; Zinder, M.; Samach, A.; Eshed, Y.; Ori, N. The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 2009, 136, 823–832.
More
Information
Subjects:
Plant Sciences
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
538
Revisions:
2 times
(View History)
Update Date:
22 Dec 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




