
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Francisco Guillén-Chable | + 2515 word(s) | 2515 | 2021-12-08 04:24:55 | | | |
| 2 | Lindsay Dong | + 406 word(s) | 2921 | 2021-12-14 03:58:25 | | |
Video Upload Options
The process of phase separation allows for the establishment and formation of subcompartmentalized structures, thus enabling cells to perform simultaneous processes with precise organization and low energy requirements. Chemical modifications of proteins, RNA, and lipids alter the molecular environment facilitating enzymatic reactions at higher concentrations in particular regions of the cell.
1. Introduction
2. Basic Thermodynamics of LLPS Driving Nucleolus Assembly
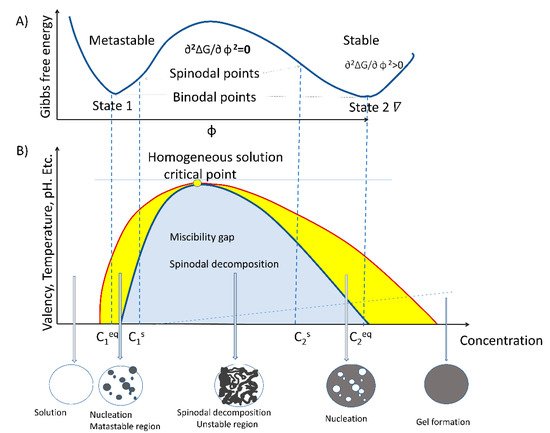
The process of LLPS is an entropy-driven event and, therefore, described by the thermodynamic law of free energy state [18][19]. Entropy-driven events are defined as spontaneous processes taking place without consuming energy. As stated by the second law of thermodynamics, spontaneous reactions occurring inside the cell take place by lowering the free energy state in pressure and temperature constants. The formation of liquid-like droplets is a passive process that involves weak intra- and inter-molecular interactions between RNA, proteins, and lipids within a dynamic scaffold [2][3][20][21][22]. LLPS-assembled biocondensates typically display an almost spherical shape, propensity for fusion and/or fission, and a high degree of internal molecule dynamics [23][24]. Moreover, LLPS is required for processes related to rRNA transcription [11], mRNA metabolism [25], posttranslational modifications (PTM) of proteins, and signaling. For example, nucleolar structure formation by coalescence assembly involves two major driving forces: a passive thermodynamically-dependent process and an active energy-consuming process [26][27]. The two specific features of the droplet formation model by coalescence are temperature-dependence and reversibility. LLPS is a spontaneous mechanism that builds up the nucleolar region and possibly mediates the functional formation of a nucleolus by spinodal decomposition primarily in the free Gibbs energy. The process of spinodal decomposition involves a thermodynamically unstable solution that can transform within a miscibility gap to a mixture of two phases, since it occurs in a thermodynamically unstable state. The spinodal region of the phase diagram for each compound is where the free energy can be lowered by allowing the components to separate. Therefore, this results in an increased relative concentration of a component material (RNA, protein, or lipid) in a particular region of the cell. The concentration will continue to increase, thus, forming a particular structure (nucleoli, Cajal body, etc.). Once nucleation begins, a structure increases in size. To maintain the dynamics of the molecular characteristics of proteins, RNA and lipids must change by chemical modifications. Phosphorylation and methylation are some of the most common chemical alterations of these three types of molecules.
Very large regions of material will change their concentration slowly due to the amount of material that must move following passive processes, from high to low gradient concentration, isoelectric point, and electrostatic gradients. Very small regions composed of just a few molecules will shrink away due to the energy cost in entropy to maintain an interface between two dissimilar component materials [28][29][30]. It does not require nucleation events to form as the phases evolve continuously, thus, creating a separation of molecules without the aid of chemical energy. However, it is interesting to note that not only does LLPS mediate the formation of the nucleolus, but energy-consuming processes are also involved. Therefore, the enzymatic process is the second force that contributes to the formation of a functional nucleolus [17]. LLPS is dependent on the multivalent crosslinking interactions of molecules, such as RNA and proteins, that possess unstructured metastable regions or intrinsically disordered regions (IDR) [8][31]. These structural features give rise to typical properties of membraneless biocondensates, such as viscoelasticity and the fusion dependent on temperature [28][29][30]. Defining the energy required in the unmixing binary or ternary mixtures is not the only phase occurrence where energy-free growth occurs. Consider an example of two independent protein–lipid complexes, which undergo a particular order structure in thermal equilibrium. The phase transition forms a disordered state, where the two molecule species A and B are randomly distributed in the nucleolus forming an ordered arrangement depending on their concentration, thus creating a particular phase as described earlier by theoretical spinodal order, [32]. Phase separation of proteins that interact with nuclear lipids appear to form a pattern structure where each molecule occupies a defined position; therefore, the diffusion would follow a similar pattern as a spinodal decomposition.
However, nucleation also plays a role, under different concentrations for some of the compounds. In addition to the complexity of the mixture of materials, we have to consider the additional complexity of molecule modification by phosphorylation, acetylation, methylation, etc. [33]. All this alteration may allow a particular molecule to change its physicochemical characteristics, and since it would be at a different concentration, it may change position as it moves from a spinodal to a binodal or outside the range of phase separation. Thus, a molecule can transit from a particular structure to another without the addition of chemical energy. Figure 1 shows the miscibility gap where the initial free energy G would allow for a particular molecule of C1 concentration to phase separate from solution. The unstable zone where the spinodal decomposition lies is where the tangent of the curve from the Gibbs free energy changes so that the ∂2G/∂ Φ 2 = 0. Therefore, molecules can phase separate freely in this region and form particular structures without additional energy. In the cell nucleus, there is an added complexity of molecules, such as RNA, that may form nucleation events that can influence the range of concentrations for phase separation. Moreover, some of the molecules may have “fixed” positions due to interactions with specific DNA sequences. Nucleation requires additional energy, as stated in ∂2∆G/∂ φ 2 > 0. This may range from lncRNA to DNA binding proteins to set the particular locations for nucleation to take place. LncRNA may provide an additional dynamic situation. Previous studies have shown that RNA can alter the concentration required for phase separation for particular proteins [34][35]. Furthermore, the addition of RNA inhibitors, such as actinomycin D or DRB, result in nucleolar alteration and form different phase separations, forming the well-known nucleolar CAP [36][37]. Therefore, the range of concentrations in which a molecule can freely form a particular phase may be wider in the presence of RNA than if the compounds were taken in an in vitro scenario. These dynamics may prove relevant for nuclear organization, in particular, during the cell cycle as well as during temperature stress, where particular proteins, such as fibrillarin, that participate in the nucleolar process may redistribute to the cytoplasm [38].
3. The Membraneless Nucleolar Compartment
4. Intrinsically Disordered Proteins Promote LLPS in the Nucleolus
4.1. Intrinsically Disordered Proteins and Intrisnically Disordered Regions
4.2. Physiochemical Features and Sequences Signatures in IDR-Containing Proteins
4.3. Posttranslational Modifications Regulating IDR-Containing Proteins
4.4. Key Nucleolar Proteins Containing IDR
References
- Heald, R.; Cohen-Fix, O. Morphology and function of membrane-bound organelles. Curr. Opin. Cell Biol. 2014, 26, 79–86.
- Banani, S.F.; Rice, A.M.; Peeples, W.B.; Lin, Y.; Jain, S.; Parker, R.; Rosen, M.K. Compositional Control of Phase-Separated Cellular Bodies. Cell 2016, 166, 651–663.
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298.
- Falahati, H.; Haji-Akbari, A. Thermodynamically driven assemblies and liquid–liquid phase separations in biology. Soft Matter 2019, 15, 1135–1154.
- Riback, J.A.; Zhu, L.; Ferrolino, M.C.; Tolbert, M.; Mitrea, D.M.; Sanders, D.W.; Wei, M.-T.; Kriwacki, R.W.; Brangwynne, C.P. Composition-dependent thermodynamics of intracellular phase separation. Nature 2020, 581, 209–214.
- Strom, A.R.; Brangwynne, C.P. The liquid nucleome—Phase transitions in the nucleus at a glance. J. Cell Sci. 2019, 132, jcs235093.
- Zhu, L.; Richardson, T.M.; Wacheul, L.; Wei, M.-T.T.; Feric, M.; Whitney, G.; Lafontaine, D.L.J.J.; Brangwynne, C.P. Controlling the material properties and rRNA processing function of the nucleolus using light. Proc. Natl. Acad. Sci. USA 2019, 116, 17330–17335.
- Castano, E.; Yildirim, S.; Fáberová, V.; Krausová, A.; Uličná, L.; Paprčková, D.; Sztacho, M.; Hozák, P. Nuclear Phosphoinositides-Versatile Regulators of Genome Functions. Cells 2019, 8, 649.
- Latonen, L. Phase-to-Phase with Nucleoli—Stress Responses, Protein Aggregation and Novel Roles of RNA. Front. Cell. Neurosci. 2019, 13, 151.
- Alshareedah, I.; Moosa, M.M.; Raju, M.; Potoyan, D.A.; Banerjee, P.R. Phase transition of RNA−protein complexes into ordered hollow condensates. Proc. Natl. Acad. Sci. USA 2020, 117, 15650–15658.
- Correll, C.C.; Bartek, J.; Dundr, M. The Nucleolus: A Multiphase Condensate Balancing Ribosome Synthesis and Translational Capacity in Health, Aging and Ribosomopathies. Cells 2019, 8, 869.
- Berry, J.; Weber, S.C.; Vaidya, N.; Haataja, M.; Brangwynne, C.P. RNA transcription modulates phase transition-driven nuclear body assembly. Proc. Natl. Acad. Sci. USA 2015, 112, E5237–E5245.
- Yang, K.; Yang, J.; Yi, J. Nucleolar Stress: Hallmarks, sensing mechanism and diseases. Cell Stress 2018, 2, 125–140.
- Hernandez-Verdun, D. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 2011, 2, 189–194.
- Bersaglieri, C.; Santoro, R. Genome Organization in and around the Nucleolus. Cells 2019, 8, 579.
- Larson, A.G.; Elnatan, D.; Keenen, M.M.; Trnka, M.J.; Johnston, J.B.; Burlingame, A.L.; Agard, D.A.; Redding, S.; Narlikar, G.J. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 2017, 547, 236–240.
- Wei, M.T.; Chang, Y.C.; Shimobayashi, S.F.; Shin, Y.; Strom, A.R.; Brangwynne, C.P. Nucleated transcriptional condensates amplify gene expression. Nat. Cell Biol. 2020, 22, 1187–1196.
- Brangwynne, C.P. Phase transitions and size scaling of membrane-less organelles. J. Cell Biol. 2013, 203, 875–881.
- Lafontaine, D.L.J. Birth of Nucleolar Compartments: Phase Separation-Driven Ribosomal RNA Sorting and Processing. Mol. Cell 2019, 76, 694–696.
- Nozawa, R.-S.S.; Yamamoto, T.; Takahashi, M.; Tachiwana, H.; Maruyama, R.; Hirota, T.; Saitoh, N. Nuclear microenvironment in cancer: Control through liquid-liquid phase separation. Cancer Sci. 2020, 111, 3155–3163.
- Lin, Y.; Currie, S.L.; Rosen, M.K. Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J. Biol. Chem. 2017, 292, 19110–19120.
- Brangwynne, C.P.; Mitchison, T.J.; Hyman, A.A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA 2011, 108, 4334–4339.
- Weber, S.C.; Brangwynne, C.P. Getting RNA and Protein in Phase. Cell 2012, 149, 1188–1191.
- Cho, W.-K.; Spille, J.-H.; Hecht, M.; Lee, C.; Li, C.; Grube, V.; Cisse, I.I. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 2018, 361, 412–415.
- Falahati, H.; Pelham-Webb, B.; Blythe, S.; Wieschaus, E. Nucleation by rRNA Dictates the Precision of Nucleolus Assembly. Curr. Biol. 2016, 26, 277–285.
- Falahati, H.; Wieschaus, E. Independent active and thermodynamic processes govern the nucleolus assembly in vivo. Proc. Natl. Acad. Sci. USA 2017, 114, 1335–1340.
- Li, P.; Banjade, S.; Cheng, H.-C.C.; Kim, S.; Chen, B.; Guo, L.; Llaguno, M.; Hollingsworth, J.V.; King, D.S.; Banani, S.F.; et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 2012, 483, 336–340.
- Yoshizawa, T.; Nozawa, R.-S.S.; Jia, T.Z.; Saio, T.; Mori, E. Biological phase separation: Cell biology meets biophysics. Biophys. Rev. 2020, 12, 519–539.
- Caragine, C.M.; Haley, S.C.; Zidovska, A. Surface Fluctuations and Coalescence of Nucleolar Droplets in the Human Cell Nucleus. Phys. Rev. Lett. 2018, 121, 148101.
- Deiana, A.; Forcelloni, S.; Porrello, A.; Giansanti, A. Intrinsically disordered proteins and structured proteins with intrinsically disordered regions have different functional roles in the cell. PLoS ONE 2019, 14, e0217889.
- Wagner, R.; Kampmann, R.; Voorhees, P.W. Homogeneous Second-Phase Precipitation. Phase Transform. Mater. 2001, 5, 309–407.
- Heinkel, F.; Abraham, L.; Ko, M.; Chao, J.; Bach, H.; Hui, L.T.; Li, H.; Zhu, M.; Ling, Y.M.; Rogalski, J.C.; et al. Phase separation and clustering of an ABC transporter in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2019, 116, 16326–16331.
- Weber, S.C. Evidence for and against Liquid-Liquid Phase Separation in the Nucleus. Non-Coding RNA 2019, 5, 50.
- Sanders, D.W.; Kedersha, N.; Lee, D.S.W.; Strom, A.R.; Drake, V.; Riback, J.A.; Bracha, D.; Eeftens, J.M.; Iwanicki, A.; Wang, A.; et al. Competing Protein-RNA Interaction Networks Control Multiphase Intracellular Organization. Cell 2020, 181, 306–324.e28.
- Sobol, M.; Yildirim, S.; Philimonenko, V.V.; Marášek, P.; Castaño, E.; Hozák, P. UBF complexes with phosphatidylinositol 4,5-bisphosphate in nucleolar organizer regions regardless of ongoing RNA polymerase I activity. Nucleus 2013, 4, 478–486.
- Elbaum-Garfinkle, S.; Kim, Y.; Szczepaniak, K.; Chen, C.C.-H.; Eckmann, C.R.; Myong, S.; Brangwynne, C.P. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. USA 2015, 112, 7189–7194.
- Liu, Y.; Liang, S.; Tartakoff, A.M. Heat shock disassembles the nucleolus and inhibits nuclear protein import and poly(A)+ RNA export. EMBO J. 1996, 15, 6750–6757.
- Cahn, J.W.; Hilliard, J.E. Free Energy of a Nonuniform System. I. Interfacial Free Energy. J. Chem. Phys. 1958, 28, 258–267.
- Nazar, R.N. Ribosomal RNA processing and ribosome biogenesis in eukaryotes. IUBMB Life 2004, 56, 457–465.
- Emmott, E.; Hiscox, J.A. Nucleolar targeting: The hub of the matter. EMBO Rep. 2009, 10, 231–238.
- Stenström, L.; Mahdessian, D.; Gnann, C.; Cesnik, A.J.; Ouyang, W.; Leonetti, M.D.; Uhlén, M.; Cuylen-Haering, S.; Thul, P.J.; Lundberg, E. Mapping the nucleolar proteome reveals a spatiotemporal organization related to intrinsic protein disorder. Mol. Syst. Biol. 2020, 16, e9469.
- Rodriguez-Corona, U.; Sobol, M.; Rodriguez-Zapata, L.C.; Hozak, P.; Castano, E. Fibrillarin from Archaea to Human. Biol. Cell 2015, 107, 159–174.
- Masuzawa, T.; Oyoshi, T. Roles of the RGG Domain and RNA Recognition Motif of Nucleolin in G-Quadruplex Stabilization. ACS Omega 2020, 5, 5202–5208.
- Hofweber, M.; Dormann, D. Friend or foe-post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 2019, 294, 7137–7150.
- Pontvianne, F.; Abou-Ellail, M.; Douet, J.; Comella, P.; Matia, I.; Chandrasekhara, C.; Debures, A.; Blevins, T.; Cooke, R.; Medina, F.J.; et al. Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet. 2010, 6, e1001225.
- Lapeyre, B.; Bourbon, H.; Amalric, F. Nucleolin, the major nucleolar protein of growing eukaryotic cells: An unusual protein structure revealed by the nucleotide sequence. Proc. Natl. Acad. Sci. USA 1987, 84, 1472–1476.
- Serin, G.; Joseph, G.; Ghisolfi, L.; Bauzan, M.; Erard, M.; Amalric, F.; Bouvet, P. Two RNA-binding Domains Determine the RNA-binding Specificity of Nucleolin. J. Biol. Chem. 1997, 272, 13109–13116.
- Girard, J.P.; Lehtonen, H.; Caizergues-Ferrer, M.; Amalric, F.; Tollervey, D.; Lapeyre, B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 1992, 11, 673–682.
- Chong, P.A.; Vernon, R.M.; Forman-Kay, J.D. RGG/RG Motif Regions in RNA Binding and Phase Separation. J. Mol. Biol. 2018, 430, 4650–4665.
- Maiser, A.; Dillinger, S.; Längst, G.; Schermelleh, L.; Leonhardt, H.; Németh, A. Super-resolution in situ analysis of active ribosomal DNA chromatin organization in the nucleolus. Sci. Rep. 2020, 10, 7462.
- Rodriguez-Corona, U.; Pereira-Santana, A.; Sobol, M.; Rodriguez-Zapata, L.C.; Hozak, P.; Castano, E. Novel Ribonuclease Activity Differs between Fibrillarins from Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 1878.
- Mitrea, D.M.; Cika, J.A.; Guy, C.S.; Ban, D.; Banerjee, P.R.; Stanley, C.B.; Nourse, A.; Deniz, A.A.; Kriwacki, R.W. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife 2016, 5, e13571.
- Bah, A.; Vernon, R.M.; Siddiqui, Z.; Krzeminski, M.; Muhandiram, R.; Zhao, C.; Sonenberg, N.; Kay, L.E.; Forman-Kay, J.D. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature 2014, 519, 106–109.
- Gibbs, E.; Perrone, B.; Hassan, A.; Kümmerle, R.; Kriwacki, R. NPM1 exhibits structural and dynamic heterogeneity upon phase separation with the p14ARF tumor suppressor. J. Magn. Reson. 2020, 310, 106646.
- Yildirim, S.; Castano, E.; Sobol, M.; Philimonenko, V.V.; Dzijak, R.; Venit, T.; Hozák, P. Involvement of phosphatidylinositol 4,5-bisphosphate in RNA polymerase I transcription. J. Cell Sci. 2013, 126, 2730–2739.
- Snead, W.T.; Gladfelter, A.S. The Control Centers of Biomolecular Phase Separation: How Membrane Surfaces, PTMs, and Active Processes Regulate Condensation. Mol. Cell 2019, 76, 295–305.
- Hoboth, P.; Sztacho, M.; Šebesta, O.; Schätz, M.; Castano, E.; Hozák, P. Nanoscale mapping of nuclear phosphatidylinositol phosphate landscape by dual-color dSTORM. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158890.
- Tran, E.J.; Zhang, X.; Maxwell, E.S. Efficient RNA 2’-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C’/D’ RNPs. EMBO J. 2003, 22, 3930–3940.
- Dragon, F.; Gallagher, J.E.G.; Compagnone-Post, P.A.; Mitchell, B.M.; Porwancher, K.A.; Wehner, K.A.; Wormsley, S.; Settlage, R.E.; Shabanowitz, J.; Osheim, Y.; et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 2002, 417, 967–970.
- Watkins, N.J.; Bohnsack, M.T. The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA 2012, 3, 397–414.
- Yin, Q.F.; Yang, L.; Zhang, Y.; Xiang, J.F.; Wu, Y.W.; Carmichael, G.G.; Chen, L.L. Long Noncoding RNAs with snoRNA Ends. Mol. Cell 2012, 48, 219–230.
- Smith, D.-L.; Erce, M.A.; Lai, Y.-W.; Tomasetig, F.; Hart-Smith, G.; Hamey, J.J.; Wilkins, M.R. Crosstalk of phosphorylation and arginine methylation in disordered SRGG repeats of S. cerevisiae fibrillarin and its association with nucleolar localisation. J. Mol. Biol. 2019, 432, 448–466.
- Martin, E.W.; Holehouse, A.S. Intrinsically disordered protein regions and phase separation: Sequence determinants of assembly or lack thereof. Emerg. Top. Life Sci. 2020, 4, 307–329.
- Protter, D.S.W.W.; Rao, B.S.; Van Treeck, B.; Lin, Y.; Mizoue, L.; Rosen, M.K.; Parker, R. Intrinsically Disordered Regions Can Contribute Promiscuous Interactions to RNP Granule Assembly. Cell Rep. 2018, 22, 1401–1412.
- Pecoraro, A.; Carotenuto, P.; Franco, B.; De Cegli, R.; Russo, G.; Russo, A. Role of uL3 in the Crosstalk between Nucleolar Stress and Autophagy in Colon Cancer Cells. Int. J. Mol. Sci. 2020, 21, 2143.




