
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mariaimmacolata Preiano' | + 8268 word(s) | 8268 | 2021-12-06 02:45:10 | | | |
| 2 | Camila Xu | -28 word(s) | 8240 | 2021-12-13 04:37:46 | | |
Video Upload Options
The urgent need to fight the COVID-19 pandemic has impressively stimulated the efforts of the international scientific community, providing an extraordinary wealth of studies. Improving the current laboratory testing methods and developing new rapid and reliable diagnostic approaches might be useful in managing contact tracing in the fight against both the original SARS-CoV-2 strain and the new, potentially fast-spreading CoV-2 variants. Mass Spectrometry (MS)-based testing methods and more specifically MALDI-MS have demonstrated without any doubt the great potential to overcome many unresolved analytical challenges arising from currently used laboratory testing assays, becoming an effective proteomic tool in several applications, including pathogen identification. With the aim of highlighting the challenges and opportunities that derive from MALDI-based approaches for the detection of SARS-CoV-2 and its variants, we extensively examined the most promising proofs of concept for MALDI studies related to the COVID-19 outbreak.
1. Introduction
However, RT-qPCR may suffer from the lack of sensitivity for the detection of SARS-CoV-2 in the early stages of infection as the concentration level of the virus is low in the upper respiratory airways during the first 6–8 days of illness, only reaching the peak in a window of 10–14 days from the onset of the illness [7][8]. Other limitations of the RT-qPCR assay are its low-throughput capacity due to many intermediate steps and long turnaround times, and the use of a poorly designed specimen extraction control [9][10]. Additionally, false-positive and high false-negative rates ranging from 1% to 30% may arise, depending on several factors such as improper sampling, lack of specificity due to poor sensitivity, and cross- and carryover contamination [11]. All together, these factors might lead to misdiagnosis and the spread of infection.
Concerning the rapid antigen tests, these are also being widely adopted for preliminary screening, but they have substantially lower sensitivity than the WHO-recommended standard, especially for pre-symptomatic and asymptomatic cases [12].
Thus, it is of crucial importance in this particular circumstance to develop novel, robust laboratory diagnostic tests that target SARS-CoV-2 with a high level of reproducibility, specificity, and sensitivity to support already existing tests.
Owing to the huge amount of data that can be rapidly collected and considerable technological advancements, mass spectrometry (MS)-based omic approaches have demonstrated, without any doubt, the possibility of performing extensive and sensitive analyses, not only opening new opportunities for the diagnosis and assessment of disease progression, but also providing insights into mechanisms of the disease[13][14].
Additionally, the use of MS platform technologies has already been applied in disease outbreaks for the study and analysis of infectious disease agents such as viruses and bacteria[15][16][17][18], suggesting its potential as the new alternative and highly specific test for the SARS-CoV-2 infection.
In particular, recent reviews confirm MS-based omic technologies have greatly contributed to the detection of SARS-CoV-2 [14][19][20].
Many efforts have been explored to make the MS-detection process high-throughput, highly accurate, and sensitive [21][22][23].
In this scenario, Matrix-Assisted-Laser-Desorption/Ionization (MALDI)-MS seems to satisfy particularly the high-throughput requirements as well as the rapid data acquisition [24]. In addition, the advancements reached by the Fourier-Transform Ion Cyclotron Resonance (FT-ICR) technology in terms of high and ultra-high-resolution power and mass accuracy in modern mass instruments make possible the development of high-throughput, highly accurate, and highly sensitive processes [25].
MALDI-MS has become one of the main proteomic tools for its relatively high tolerance of mixtures and biological contaminants [24]. This technique is largely used for the characterization of biomolecules, especially proteins and peptides. However, the original purpose of this technology has also been redirected to microorganism identification. In the last two decades, MALDI has been successfully integrated into the microbiology laboratory workflow due to its capacity for rapid, low-cost microbial species identification, and it is now routinely adopted in Europe and the United States for bacteria identification. Therefore, considering that most clinical diagnostic laboratories already have MALDI-MS equipment, its application in emerging pathogen research as a screening method for COVID-19 detection in large populations presents great potential.2. MALDI-TOF and MALDI-FT-ICR Mass Spectrometry: A Brief Presentation
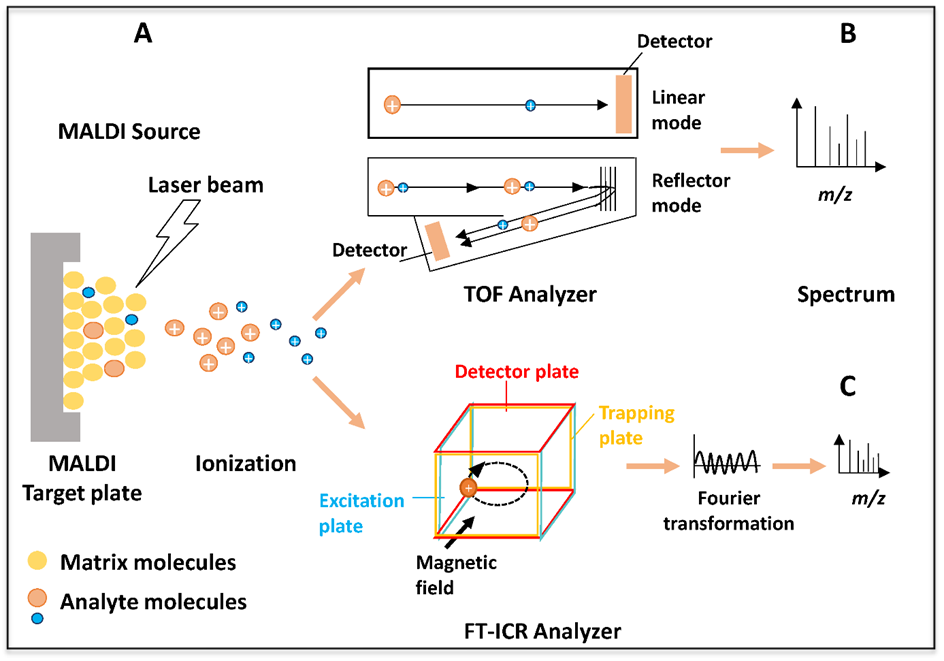
As also illustrated in Figure 1B, in modern MALDI-TOF instruments, sample analysis can be performed using linear and/or reflector MS modes. The linear MS mode is well-suited for analytes with molecular weights above 4000 Da and enables very sensitive analyses. Analytes below 4000 Da are analyzed in reflector MS mode, with improved mass accuracy and resolution. Specifically, when operating in reflector MS mode, a reflector mirror which reflects ions back toward the reflector detector is used. Therefore, a longer flight ion path is provided, which increases ion separation through an enhanced resolution (Figure 1B). The reflector (also used in the MS/MS mode) is a two-stage electrostatic mirror consisting of a series of plates. The reflector also improves mass accuracy and resolution by correcting the dispersion time caused by variations of initial kinetic energy that generate slight differences in ion velocity and by filtering out neutral molecules. In the presence of a second TOF analyzer (MALDI-TOF/TOF), samples analyzed in the reflector MS mode may be further analyzed by the MS/MS mode to obtain a fragmentation pattern used to determine structural information. This platform is especially used in peptide/protein identification and amino acid sequencing. MALDI-TOF MS allows for the analysis of hundreds of proteins or other biomolecules in a single spectrum and has been used in several diagnostic applications in recent years, including: (i) the study of the distributions of particular biochemical compounds on tissues (MALDI-based imaging MS)[29][30];(ii) the analysis of nucleic acids for genotyping single nucleotide polymorphisms (SNPs) [31](iii) the profiling of biological samples to aid in the discovery of new of disease biomarkers in exploratory studies with diagnostic applicability. Such biomarkers might be used as indicators of progress or the severity of diseases/disorders[32][33][34][35][36][37], and for (iv) the identification, characterization, and typing of bacteria, yeasts, and viruses [38][39]. Although less used than TOF because of its high costs, another mass analyzer coupled with a MALDI source is the FT-ICR, which provides better performance than a MALDI-TOF instrument in terms of resolution, mass accuracy, and dynamic range. The FT-ICR analyzer measures the mass-to-charge ratio (m/z) of ions according to the cyclotron frequency of the ions (Figure 1C). After ion generation in the MALDI source, the ions are transferred to the Penning trap consisting of a cell under a high vacuum positioned in a superconducting magnet. The ions are excited by a radiofrequency and generate a signal. The frequency of the signal from each ion is equal to its orbital frequency, which is inversely related to its m/z value. The signal intensity of each frequency is proportional to the number of ions with that m/z value. The signal is amplified and all the frequency components are determined. The mass spectrum is generated using Fourier analysis (Figure 1C). The frequencies match the m/z, and the amplitudes are correlated with the abundance of the analytes [40]. In comparison to any other mass analyzer, FT-ICR provides the highest mass accuracy and mass resolving power, reaching up to ppb mass accuracy, high dynamic range, and mass resolving power greater than 1,000,000 in routine analyses. Very recently, molecular structures with m/z differences of 1.79 mDa were resolved and identified with high mass accuracy and sensitivity [26]. For these reasons, MALDI-FT-ICR is very suitable for the analysis of complex biological samples in proteomic, metabolomic, and lipidomic research in different fields of application, including microbiology, imaging, and biomarker discovery[41][42].
3. MALDI-MS for Pathogen Detection: A General Overview
|
Specimen |
Sample Size |
MS Instrumentation |
Identification Method |
Molecular Target |
Use of Database or Algorithms for Identification |
Time |
Diagnostic Performances |
References |
|
Nasopharyngeal samples |
Not specified |
MALDI-FT-ICR |
Proteotyping |
Viral proteins: - N (1); - S (2); - M (3). |
Online database searching |
Similar to RT-PCR time frame (few minutes for mass spectra acquisition) |
Only analytical performances |
Dollman et al. |
|
Saliva or gargle samples |
35 samples |
MALDI-TOF MS |
Biotyping and Biomolecular Host Profiling |
Host proteins; viral proteins: - S; - Veps (4). |
Output data processed with appropriate software (not specified) |
45 min for sample preparation, 3 min per sample for MALDI-TOF analysis, for data results analysis |
Sensitivity of ~100% (5) |
Iles et al. |
|
Gargle samples |
- 30 COVID-19 positive samples (89% asymptomatic) - 30 COVID-19 negative samples |
MALDI-TOF MS |
Biotyping and Biomolecular Host Profiling |
Host proteins; viral proteins: - S. |
Online database searching |
Not mentioned |
- Sensitivity of 93.33–100% - Specificity of 90–93.33% (6) |
Chivte et al. |
|
Oral or nasopharyngeal samples |
- 22 COVID-19 positive samples - 22 COVID-19 negative samples |
MALDI-TOF MS |
Genotyping |
Viral genes: - N1; - N2; - N3; - ORF1 (7); - ORF1ab. |
Not specified |
8 h for the entire process |
Not mentioned |
Wandernoth et al. |
|
Oral or nasopharyngeal samples |
168 suspected COVID-19 samples |
MALDI-TOF MS |
Genotyping |
Viral genes: - N1; - N2; - N3; - ORF1; - ORF1ab. |
Online database searching |
8 h for the entire process |
Not mentioned |
Rybicka et al. |
|
Saliva samples |
- 34 COVID-19 positive samples - 26 COVID-19 negative samples |
MALDI-TOF MS |
Genotyping |
Viral genes: - N1; - N2; - N3; - ORF1; - Orf1ab. |
Not specified |
Not mentioned |
- Sensitivity of 97.14% - Specificity of 100% (8) |
Hernandez et al. |
|
Nasopharyngeal samples |
- 211 COVID-19 positive samples - 151 COVID-19 negative samples |
MALDI-TOF MS |
Biomolecular Host Profiling |
Host proteins |
Machine learning algorithms |
Not mentioned |
- Sensitivity of 94.7% - Specificity of 92.6% (9) |
Nachtigall et al. |
|
Nasal samples |
- 107 COVID-19 positive samples (28 asymptomatic and 79 symptomatic) - 92 COVID-19 negative samples |
MALDI-TOF MS |
Biomolecular Host Profiling |
Host proteins |
Machine learning algorithms |
Total turnaround time < 1 h |
- Accuracy of 98.3%, PPA (10) of 100%, NPA (11) of 96% for DNN (12) model - Accuracy of 96.6%, PPA of 98.5%, and NPA of 94% for GBM (13) model (9) |
Tran et al. |
|
Nasopharyngeal samples |
237 samples |
MALDI-TOF MS |
Biomolecular Host Profiling |
Host proteins |
Machine learning algorithms |
Turnaround time < 2 h |
- Sensitivity of 100% - Specificity of 92% - Accuracy of 97% (9) |
Deulofeu et al. |
|
Nasopharyngeal samples |
311 samples |
MALDI-TOF MS |
Biomolecular Host Profiling |
Host proteins |
In-house database searching and machine learning algorithms |
Not mentioned |
- Sensitivity of 61.76% - Specificity of 71.72% - Accuracy of 67.66% (9) |
Rocca et al. |
|
Serum samples |
- 146 COVID-19 positive samples -152 COVID-19 negative samples |
MALDI-TOF MS |
Biomolecular Host Profiling |
Host proteins |
Machine learning algorithms |
Less than 1 min per sample for MALDI-TOF analysis |
- Sensitivity of 98% - Specificity of 100% - Accuracy of 99% (9) |
Yan et al. |
(1) N: nucleocapsid protein; (2) S: spike protein; (3) M: membrane protein; (4) Veps: viral envelope proteins; (5) Diagnostic sensitivity of the method is reported; (6) Sensitivity and specificity are referred to AUC and ROC curve analyses; (7) ORF: open reading frame; (8) Percent agreement and Cohen’s kappa were calculated to assess sensitivity and specificity; (9) Diagnostic performances are referred to the Machine learning model used. (10) PPA: positive percent agreement; (11) NPA: negative percent agreement; (12) DNN: deep neural network; (13) GBM: XGBoost gradient boosting machine.
4. MALDI-MS Investigations Targeting SARS-CoV-2
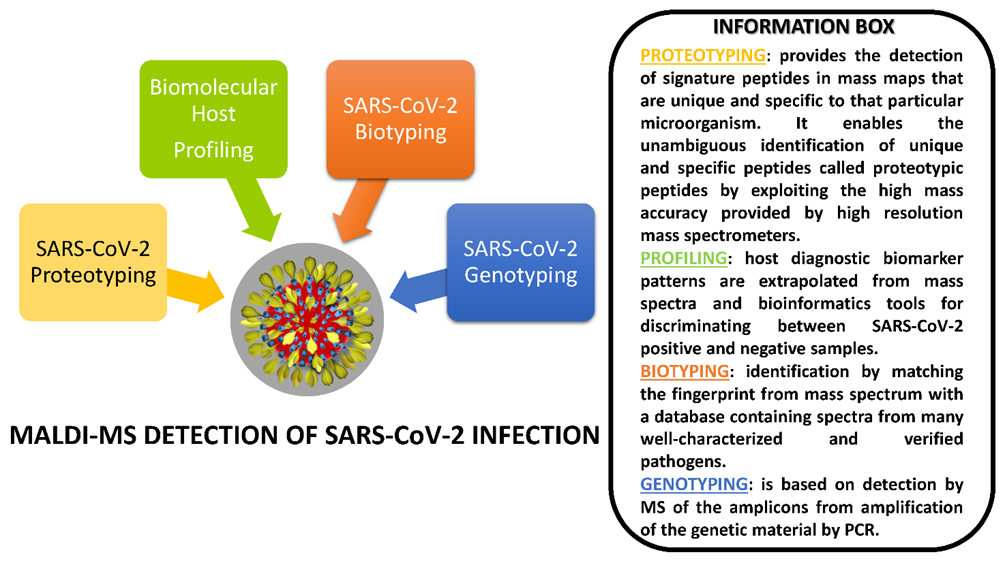
In this section, we report the MALDI-MS-based investigations which aimed to detect SARS-CoV-2 infection with the use of “proteotyping” [52], “biotyping” [47][48], “genotyping”[49][50][51], and biomolecular host profiling[53][54][55][56][57] approaches. Interestingly, two of these investigations used both biotyping and biomolecular host profiling strategies[47][48]. All these investigations were extensively reviewed in a very recently published paper [65].
4.1. Proteotyping Approach for SARS-CoV-2 Detection by MALDI-MS
Dollman and colleagues applied a proteotyping strategy for the detection of peptides unique to the SARS-CoV-2 by mass alone; they used High Resolution MALDI-FT-ICR Mass Spectrometry[52]. In particular, nasopharyngeal (NP) swabs from infected patients were cultured in Vero E6 cells or directly prepared before the MALDI-MS analysis. The use of a virus culture provided a more reliable approach to overcome potential false negatives deriving from specimens with low viral loads. The authors performed the MALDI-FT-ICR-MS analyses on the . In the case of cell-cultured SARS-CoV-2 samples with higher virus titers in comparison to other samples, twenty of the most prominent signals were confidently matched to fragments of the N, M, and S proteins with a high mass accuracy (<3 ppm) (Table 1). Detected peptides covered a large range of identified N- and C-terminal, fusion peptide receptor, and RNA-binding domains.
The acquired mass spectra from the non-cell-cultured NP samples showed peptide ions with reduced intensity and lower signal-to-noise ratio (S/N) as compared to those from experiments using cell-cultured samples; in this case, additional matrix cluster ions were also observed. However, despite the lower virus titers, the tryptic peptides of M, N, and S proteins were detected, including a new peptide at m/z 843.4255. In particular, six peptides were always detected in ten different NP samples, and five represent reliable signature peptides for SARS-CoV-2 proteotyping. In fact, due to the shared clinical symptoms presented by the SARS-CoV-2 with other circulating influenza viruses, the authors, by means of a specifically constructed algorithm and database, showed that these peptides were detected in all SARS-CoV-2 strains and exhibited a difference in mass greater than 3 ppm in comparison to those fragments detected in more than 95% of the cases from all influenza virus proteins of the circulating strains.
Table 2 reports the limit of detection (LOD) of the various analytical assays (including LOD related to conventional RT-PCR [66]) calculated based on the information from each of the authors, in order to make the comparison more straightforward for the readers. To determine the LOD of this MS platform, serial dilution experiments on the cell-cultured sample were performed, and the most abundant viral peptide signal at m/z 1635.8238 was monitored by MS. The peak was detected down to 0.75 ng, matching 7.5 × 105 copies (Table 2).
This represents an excellent result, considering that a recent study reported maximum estimated levels of virus collected by NP swabs varying from 106 to 109 copies
Interestingly, the authors stressed that this copy number is comparable with the 105106 copies of virus necessary for high-quality PCR sequencing; however, as they additionally claimed, it is also higher than the 103–104 copies usually required for virus detection by PCR (Table 2). As underlined by the authors, the detection limits of 105 copies could be significantly improved when operating in an automated selected ion monitoring (SIM) strategy for the detection of only selected peptides for a proteotyping assay (Table 2).
Table 2. Diagnostic performances and estimated costs of the different tests used for SARS-CoV-2 detection.
|
Specimen |
Test Category |
LOD (1) |
Analysis Time |
Estimated Cost Per Sample |
Key Points |
References |
|
Respiratory tract specimens |
RT-PCR |
<10 copies/reactions (103–104 copies) (2) |
4–6 h |
USD 10–15 |
Current gold standard for COVID-19 diagnosis. |
Kevadiya et al. |
|
Nasopharyngeal samples |
MALDI-FT-ICR |
105 copies |
Similar to RT-PCR time frame |
USD 100 |
LOD could be improved in automated selected ion monitoring (SIM) strategy (reaching 103–104 copies). |
Dollman et al. |
|
Saliva or gargle samples |
MALDI-TOF MS |
~10–102 copies |
~50 min |
Less than USD 1 |
Specificity at 10–102 copies was reached only for the S1 protein peak. |
Iles et al. |
|
Gargle samples |
MALDI-TOF MS |
~30 copies |
Not mentioned |
Not mentioned |
MS protocol was sensitive and comparable with RT-PCR forlow viral loads. |
Chivte et al. |
|
Oral or nasopharyngeal samples |
RT-PCR/MALDI-TOF MS |
~10 copies |
8 h |
Not mentioned |
Time-to-results was faster for RT-PCR, while hands-on time was comparable between RT-PCR and MS assay techniques. |
Wandernoth et al. |
|
Oral or nasopharyngeal samples |
RT-PCR/MALDI-TOF MS |
~10 copies |
8 h |
~ EUR 10 |
The MS assay was able to detect SARS-CoV-2 in low viral load specimens. |
Rybicka et al. |
|
Saliva samples |
RT-PCR/MALDI-TOF MS |
~103 copies |
Not mentioned |
Not mentioned |
The LOD of 103 copies was obtained for the N2 target. |
Hernandez et al. |
(1) LOD: limit of detection; (2) 103–104 copies are typically required for PCR detection.
4.2. Biotyping Approach for SARS-CoV-2 Detection by MALDI-MS
With the aim of carefully and systematically analyzing all the challenging issues of an MS-based diagnostic process, Iles and colleagues developed a clinical MALDI-TOF MS assay for SARS-CoV-2 detection in a saliva or gargle solution [47]. The authors performed several experiments in order to optimize the MALDI-TOF detection of viral protein and its proteolytic peptide detection. They finally developed a clinical diagnostic protocol based on (1) the collection of gargle/saliva samples; (2) the rapid processing of the gargle samples by filtration, acetone precipitation with subsequent pellet resuspension, and protein solubilization; (3) MALDI TOF analysis; (4) output data processing by an appropriate software (see Table 1).
Interestingly, through the clinical information extrapolated by MALDI-TOF analysis, the authors demonstrated not only the possibility to detect S protein fragments, but also other peaks matching with Immunoglobulin light chains indicative of an oral upper respiratory immune response, and elevated levels of gargle/saliva IgA heavy chain peak indicative of a viral immune response.
These features reveal the diagnostic utility of MALDI-TOF MS as a powerful and economically ideal solution, with its ease of sampling and speed of analysis. However, other studies and validation tests on saliva/gargle spiked with cultured virus, or a direct comparison of the MALDI-TOF MS analysis with an RT-PCR detection of COVID-19 in clinical samples is required.
Starting from the results obtained by the study of Iles and coworkers, Chivte and colleagues analyzed gargle samples with MALDI-TOF MS to assess the presence of SARS-CoV-2 infection and compared the resulting spectra with the corresponding results of an RT-qPCR from NP swabs [48]. In particular, they analyzed 60 gargle samples (see Table 1) from volunteer student athletes, including 30 PCR positive and 30 PCR negative samples. Spectral clear differences were observed in the m/z range from 20,000 to 200,000. This extended m/z range allowed them to analyze the area under the curve (AUC) of putative viral and host proteins in order to assess the presence of SARS-CoV-2 infection. The receiver operating characteristic (ROC) analysis identified five peaks which showed highly sensitive and specific discrimination between COVID-19 positive and negative samples, including peaks which probably corresponded to the S1 and S2 fragments of the SARS-CoV-2 S protein, a potential biomarker near m/z 112,000, and peaks putatively assigned as human immunoglobulins or human α-amylase (Tables 1 and 3). For these biomarkers, an elevated concordance was achieved (90%) with the RT-PCR results, demonstrating that this methodology is a promising tool for a rapid and inexpensive COVID-19 assay.
Table 3. Sample size,protein patterns, and their expression trend in COVID-19 vs non-COVID-19 samples in Biomolecular Host Profiling studies.
|
Specimen |
Sample Size |
Patient Classification |
Protein/Peptide Identity |
m/z |
Expression (Downregulated ↓, Upregulated ↑) Against Control |
Bioinformatic Tool |
Sensitivity/ Specificity of ML Diagnostic |
References |
|
Gargle |
60 samples |
- 30 COVID-19 positive samples (89% asymptomatic) - 30 COVID-19 negative samples |
Immunoglobulin heavy chain or amylase |
55,500–59,000 |
↑ |
- AUC (1) - ROC (2) |
ML (3) not applied. |
Chivte et al. |
|
Immunoglobulin heavy chain doubly charged |
27,900–29,400 |
↑ |
||||||
|
Not identified |
~112,000 |
↑ |
||||||
|
Nasopharyngeal |
362 samples |
- 211 COVID-19 positive samples -151 COVID-19 negative samples |
Not identified |
3358 |
↓ |
- ML (SVM-R) (4) - PCA (5) - ROC |
- Sensitivity of 94.7% - Specificity of 92.6% |
Nachtigall et al. |
|
3095 |
↓ |
|||||||
|
4532 |
↑ |
|||||||
|
3337 |
↓ |
|||||||
|
3152 |
↑ |
|||||||
|
10,444 |
↓ |
|||||||
|
7612 |
↓ |
|||||||
|
Nasal |
199 samples |
- 107 COVID-19 positive samples (28 asymptomatic and 79 symptomatic) - 92 COVID-19 negative samples |
Not identified |
Not specified |
Not specified |
- ML (DNN (6) and GBM (7)) - PCA - AUC - ROC |
- Accuracy of 98.3%, PPA (8) of 100%, NPA (9) of 96% for DNN model - Accuracy of 96.6%, PPA of 98.5%, NPA of 94% for GBM model |
Tran et al. |
|
Nasopharyngeal |
237 samples |
Not mentioned |
Not identified |
Not specified |
Not specified |
- ML (SVM) - PCA |
- Sensitivity of 100% - Specificity of 92% |
Deulofeu et al. |
|
Nasopharyngeal |
311 samples |
Not mentioned |
Not identified |
3372 |
↓ |
- ML - AUC - ROC |
- Sensitivity of 61.76% - Specificity of 71.72% |
Rocca et al. |
|
3442 |
↓ |
|||||||
|
3465 |
↓ |
|||||||
|
3488 |
↓ |
|||||||
|
6347 |
↓ |
|||||||
|
10,836 |
↓ |
|||||||
|
Serum |
298 samples |
- 146 COVID-19 positive samples - 152 COVID-19 negative samples |
Not identified |
6357 |
↓ |
- ML (LR) (10) - AUC - ROC - PCA |
- Sensitivity of 98% - Specificity of 100% |
Yan et al. |
|
6654 |
↓ |
|||||||
|
6639 |
↓ |
|||||||
|
28,232 |
↓ |
|||||||
|
Platelet basic protein |
13,886 |
↓ |
||||||
|
Platelet factor 4 variant |
7614 |
↑ |
||||||
|
Hemoglobin subunit alpha |
15,123 |
↑ |
||||||
|
Hemoglobin subunit beta |
15,867 |
↑ |
||||||
|
WD repeat-containing protein |
28,091 |
↑ |
(1) Area under the receiver operating characteristic (ROC) curve; (2) Receiver operating characteristic; (3) Machine learning; (4) Support vector machine with a radial kernel; (5) Principal component analysis; (6) DNN: deep neural network; (7) GBM: XGBoost gradient boosting machine; (8) PPA: positive percent agreement; (9) NPA: negative percent agreement; (10) Logistic regression.
4.3. Genotyping Approach for SARS-CoV-2 Detection by MALDI-MS
Wandernoth and colleagues evaluated the ability of MALDI-TOF MS to detect nucleic acid from SARS-CoV-2 in biological samples and compared the results to those of rRT-PCR [49]. In particular, they analyzed oral and NP swab samples from 22 patients who tested positive and 22 patients who tested negative by RT-PCR for SARS-CoV-2 infection (see Table 1). They used the new commercially available MS-based assay ® (Agena Bioscience®, San Diego, CA, USA), a genotyping panel for the detection of SARS-CoV-2 developed by Agena Bioscience, which received the CE-IVD mark in Europe for the detection of nucleic acid from SARS-CoV-2 in respiratory specimens. This platform provides a robust route for the detection of the SARS-CoV-2 virus in human samples. The results of the rRT-PCR and MALDI-TOF MS analyses were comparable in all samples. The LOD was estimated at 10 genome copies (Table 2). Very recently, Rybicka and colleagues [50] also tested the new commercially available MS-based assay MassARRAY® SARS-CoV-2 Panel (Agena Bioscience) and compared it to the RT-PCR diagnostic test. Oral and NP swabs from 168 suspected COVID-19 patients with symptoms of respiratory infection were simultaneously processed with both assays (Table 1). Among the 168 analyzed samples, different results were obtained for 10.12% (17 samples). Using the MassARRAY® SARS-CoV-2 Panel, the authors were able to detect SARS-CoV-2 in low viral load specimens and with very few microliters of viral RNA. They pointed out that the MassARRAY® System provides automation, minimal hands-on time, and onboard data analysis, delivering easy-to-interpret data with a simple and fast workflow.
Also Hernandez and colleagues compared the already mentioned RT-PCR/MALDI-TOF MS-based assay (AGENA MASS ARRAY) with the cobas® 6800/8800 SARS-CoV2 real-time RT-PCR (Roche, Basel, Switzerland) for conventional real-time RT-PCR [51]. They collected saliva samples from 60 patients, which were previously subjected to molecular testing for SARS-CoV-2 in NP or anterior nare specimens.
Interestingly, SARS-CoV-2 detection in saliva samples by the AGENA system showed high sensitivity and specificity (Table 1) when compared to RT-PCR results from NP or anterior nare specimens. Analogue sensitivity and specificity were obtained by cobas® 6800/8800 SARS-CoV-2 real-time RT-PCR (Roche).
These data demonstrate that saliva constitutes an appropriate matrix for SARS-CoV-2 detection on the novel Agena system, with a comparable performance to the more ubiquitous real-time RT-PCR technology.
4.4. Biomolecular Host Profiling Approach for SARS-CoV-2 Detection by MALDI-MS
In this section, we report on MALDI-MS studies performed on a variety of biological specimens in which host diagnostic biomarker patterns had been extrapolated rather than direct SARS-CoV-2 detection [53][54][55][56][57]. The investigations by Iles and colleagues [47] and by Chivte and colleagues [48], which assessed the presence of both viral and host proteins, were already reported in the previous section (see Table 1 and Table 3). Many of these investigations used oral or respiratory samples [53][54][55][56],which are the commonly accepted specimens for SARS-CoV-2 detection; interestingly, one such study was conducted using serum samples to decipher the molecular changes induced by the systemic effects after infection [57]. In these investigations, MALDI-TOF readouts were processed by machine learning (ML) techniques (Table 1) to extrapolate novel or unknown diagnostic information from a mass spectra dataset. Additionally, the robustness and clinical significance of the ML models employed were assessed.
Nachtigall et al. described a method of detecting SARS-CoV-2 in NP swabs using the MALDI-TOF MS and ML analyses [53]. They proposed this approach starting from the rationale that several clinical laboratories use MALDI-MS as a high-throughput, cost-effective, and robust technology for the conventional diagnosis of pathogen infections.
They analyzed a total of 362 NP swab samples, 211 RT-PCR positives and 151 RT-PCR negatives, from subjects coming from three different countries (Argentina, Chile, and Peru) (Table 1).
The authors tested different feature selection methods and ML approaches to determine the top performing analysis strategies. Feature selection methods can be used in data pre-processing to obtain efficient data reduction, a useful step in order to identify accurate data models [96]. Through a comparison of the results obtained from RT–PCR and MALDI-TOF coupled with ML, the authors found a concordance rate that was acceptable as a clinical diagnostic approach (>80%), confirming that the MALDI-MS and ML analyses represent promising alternatives as fast screening assays for SARS-CoV-2.
Tran and colleagues described as proof-of-concept a novel, automated, ML-enhanced MALDI-TOF MS approach for analyzing nasal swabs from patients with suspected COVID-19 [54].
The authors evaluated the performance of the MALDI-TOF MS method using SARS-CoV-2 RNA PCR positive and negative samples in order to determine the accuracy, the positive percent agreement (PPA), and the negative percent agreement (NPA) of the MALDI-TOF MS method as compared to the PCR method. They collected 226 nasal swab samples that were analyzed both by RT-PCR and by MALDI-TOF MS. Normalized mass spectra were subjected to ML analysis by the Auto-ML MILO (Machine Intelligence Learning Optimizer) platform [67]. The MILO as ML platform consists in a series of algorithms, scalers, scorers, and feature selectors/transformers, which are used to generate models that are then statistically assessed to ultimately identify the best performing model for a specific purpose. The authors described their ML-enhanced MALDI-TOF approach as an attempt to address both throughput and speed limitations observed in molecular platforms; interestingly, the authors demonstrated the ability of both PCA and ML models to classify MALDI-TOF mass spectra for discriminating positive and negative COVID-19 cases.
The high-throughput performance of this platform was also demonstrated by the short time required for the analysis (total turnaround time was less than 1 h, with the potential of performing up to 1104 analyses per day, per instrument) as compared to commercial RT-PCR platforms, which need, for example, batch testing for optimal reagent use.
Another investigation that described the analysis of NP samples by MALDI-TOF MS coupled with ML was performed by Deulofeu and colleagues [55]. In particular, they used different ML approaches for the analysis of the MALDI-TOF mass spectra from 237 NP samples in order to identify a signature characterizing negative and positive samples for SARS-CoV-2 infection (Table 1). These samples were analyzed by both RT-PCR and MALDI-TOF MS. The obtained data demonstrated the utility of the developed method for the detection of SARS-CoV-2-positive samples and the simplicity, safety, rapidity, and cheapness of MALDI-TOF MS as a diagnostic test. The authors estimated the cost of MALDI-TOF MS to be 25% more than the cost of the RT-PCR analyses. They also stated that the time required to receive results is around 6 h for a conventional RT-PCR, whereas the analyses of the NP samples by MALDI-TOF MS take less than one-third of this (Table 1).
Finally, Rocca and colleagues also investigated the potential role of MALDI-TOF MS combined with ML methods in the identification and the discrimination between COVID-19 positive samples and COVID-19 negative ones [56]
They used NP swab samples from 311 patients that were analyzed by MALDI-TOF MS analysis without processing (Table 1). From the evaluation of parameters and statistical analysis, six peaks at m/z 3372, 3442, 3465, 3488, 6347, and 10,836 demonstrated a decrease or an absence of COVID-19 positive samples and were considered as potential biomarkers (Table 3).
The preliminary results demonstrated that the method still needs to be improved, and the authors stated that further studies on a larger cohort of patients, evaluating different extraction methods and improving ML algorithms, will be performed.
Yan et al. utilized a high-throughput serum peptidome profiling method based on MALDI-TOF MS for the detection of COVID-19 [57]. They analyzed serum samples from a total of 298 individuals: 146 COVID-19 patients (classified into mild, typical, severe, and critical) and 152 control patients (including 73 non-COVID-19 patients with similar clinical symptoms such as fever/cough, 33 tuberculosis patients, and 46 healthy individuals) (Table 1).
Using ML methods, twenty-five peaks were identified as the distinctive features between COVID-19 patients and control participants, with statistically significant differences (Table 3).
Starting from the observation that the symptoms of SARS-CoV-2 infected patients are frequently shared by patients affected by other respiratory infections, the authors highlighted that this could lead to the misinterpretation of results; so they emphasized the ability of this platform to discriminate COVID-19 patients from non-COVID-19 subjects with similar clinical symptoms. The authors concluded that these results also demonstrated that MALDI-TOF-based serum profiling could be a powerful tool for screening, routine surveillance, and diagnosis of COVID-19 in populations.
5. Discussion
5.1. Comparison of MALDI-MS vs RT-PCR Techniques for SARS-CoV-2 Detection: Advantages and Limitations
As a resource for new, alternative and/or complementary, rapid COVID-19 diagnostic tests, here we explored the recent developments and applications of MALDI MS-based technologies for the accurate and unambiguous SARS-CoV-2 clinical diagnosis, which may help to control the COVID-19 pandemic.
MS techniques have rapidly complemented or replaced conventional methods in clinical diagnostic laboratories and have been successfully implemented in the investigation of viruses and their pathogenesis [13],[15][16][17][18][68][69]. In particular, MALDI-MS has become an indispensable and versatile method for biochemical and clinical investigations [70][71].MALDI-MS represents one such technology, characterized by its ease of use, high-throughput capabilities, and cost effectiveness, thus becoming a routine tool in clinical microbiology laboratories and also proving to be capable of supporting clinical decision-making[24][44] .
The current gold standard technique for the molecular diagnosis of SARS-CoV-2 infection is the RT-qPCR, which allows for the analysis of thousands of samples in a single day and shows a high testing sensitivity of 95% [72] and a low limit of detection of <10 copies/reaction[73] under ideal circumstances. Nevertheless, several investigations have reported that clinically evident COVID-19 infections often go undetected by SARS-CoV-2 RT-PCR testing [74];the evidence that virus shedding can occur at undetectable levels during the very early and late stages of SARS-CoV-2 infection demonstrates that RT-PCR results should always be interpreted in a wider context. Indeed, the most efficient strategy for the diagnosis of SARS-CoV-2 infection in suspected patients should encompass a combination of SARS-CoV-2 detection by RT-PCR with clinical and epidemiologic observations (symptoms, previous exposure to virus, negative diagnostic tests for other respiratory diseases), whereas additional follow-up testing with RT-PCR should be required from patients with initially negative results and high suspicion of COVID-19.
The time required for conventional PCR can vary from about 4–6 h for sample processing (including sample preparation, RNA extraction, reverse transcription PCR, and readout of amplified DNA products), up to a few days, considering the time needed for the eventual transport of the specimen to the laboratory.
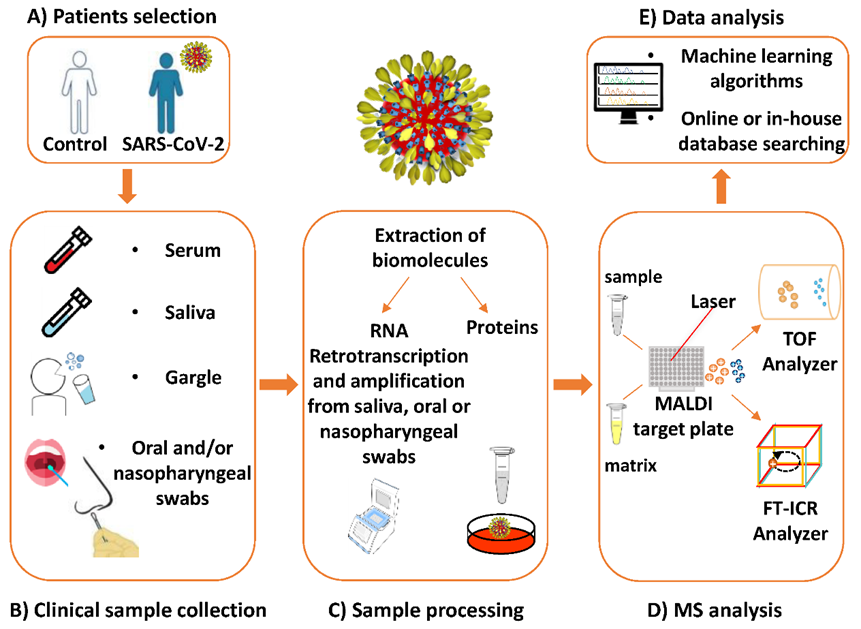
Most of the reports described here that are related to Biotyping, Proteotyping, and Genotyping approaches are proof-of-principle studies showing the feasibility of viral protein or viral nucleic acid detection by MALDI-MS, which often do not involve stress testing in low viral load samples. Nevertheless, as reported in Table 1 and Table 2, the specificity, sensitivity, and the detection limit of the SARS-CoV-2 virus by MALDI-MS diagnostic methods were high and comparable to reference methods such as PCR.
5.2. Pre-Analytical and Analytical Issues for Molecular Detection of SARS-CoV-2
There are several critical issues and challenges affecting the diagnosis of SARS-CoV-2 infections, including sampling operations, specimen source, and sampling time, which could have an impact on the end result. It is essential to handle bodily fluids according to standardized procedures in order to distinguish reliable molecular biomarkers and to assess the influence of pre-analytical parameters on the final result in order to avoid artifacts. As revealed by the literature reviewed in the previous sections, there is high variability in all of the abovementioned critical technical factors; for example, most studies do not clarify if the tested samples were analyzed by MALDI-MS immediately after collection or if they had been deep-frozen until analysis. This represents an important limitation, considering that several pre-analytical and analytical variables, including storage time and temperature, could alter the analysis of the MS profile of biological specimens, and that several studies demonstrated that the proteins could undergo degradation processes[37][75][76]. Thus, especially for the biomolecular host profiling studies, it is not possible to exclude the fact that the observed differences in the molecular fingerprints obtained by MALDI-MS could derive from the degradation of the molecular species due to the action of endo-proteases, including those of microbial origin in the nasal environment, or from the inherent biological variability among subjects, also due to genetic factors affecting the phenotype.
5.3. Detection of SARS-CoV-2 in Different Types of Clinical Specimens
Respiratory sampling is the preferred and widely approved method for SARS-CoV-2 detection by RT-qPCR. The choice of optimal clinical specimens takes into consideration several factors such as ease of accessibility, non-invasive collection procedure, a lower risk to health care professionals during sample collection, and good viral loads for higher possibilities of detection. NP and/or OP swabs are best recommended for the screening or diagnosis of an early infection, when low viral loads are known to occur and false-negative results can derive from differences in analytical sensitivity among methods[77]. For these reasons, most of the studies reviewed here are based on the use of NP or OP samples.
An alternative option for collecting upper respiratory tract specimens in order to detect SARS-CoV-2 is self-collected saliva or gargle specimen[66] . Saliva is a reliable biological fluid that can easily be provided by the patient; it does not require healthcare personnel in a screening program [66][78]. In fact, a mass screening program should rely on devices that can be used by non-medical staff to promptly assess whether an individual is positive to COVID-19. Moreover, a gargle sample is obtained from an easy-to-perform procedure and is comfortable for the patient. Reliable and rapid diagnostic techniques based on the use of saliva or gargle specimens might represent a cost-saving procedure considering that their collection does not require the need to develop and maintain specific infrastructure for swab collection as well as the need for dedicated healthcare personnel.
Serum samples could be used to understand the molecular mechanism for the diagnosis of COVID-19. As a consequence of viral infection, serum may contain mediators produced by the systemic effects or released to the lung, and this could reflect the physiological or pathological state [20].
Serum samples, and more generally, serological methods, could play a key role in the epidemiology of COVID-19 and in defining the immune status of asymptomatic patients, but they are not recommended for the screening or diagnosis of early infections [79]. However, serology could contribute to the confirmation of the diagnosis of COVID-19 by providing information about the type and the concentration of various immunoglobulins generated after SARS-CoV-2 infection [80].
5.4. Asymptomatic Infection and the Spread of New SARS-CoV-2 Variants
Another critical issue is the limited resources employed to facilitate and enable the widespread rapid screening of asymptomatic individuals necessary to control the potential of silent chains of virus transmission. Asymptomatic patients are the carriers of the live, replicating virus and contribute to the unknown viral transmission and spread. In some cases, the methods used for SARS-CoV-2 detection in these subjects can be less sensitive than in the symptomatic ones [81]. The here reviewed studies which have considered asymptomatic population demonstrated that the MALDI-TOF MS platform was able to detect SARS-CoV-2 infection in asymptomatic individuals, and with elevated accuracy (see Table 1), which could be of paramount importance for the reopening of businesses, recreational facilities, schools, and all non-hospital settings.
In addition to asymptomatic infection, SARS-CoV-2 variants are another challenge for virus detection[82][83] . In fact, new variants caused by mutations in the genome of the SARS-CoV-2 are now emerging as the virus spreads throughout the world, and this is of great relevance considering that such mutations may alter various aspects of virus biology such as pathogenicity, infectivity, transmissibility, and/or antigenicity [84]]. Thus, alternative and/or complementary methods of rapidly detecting and monitoring the evolution of virus strains are of vital importance.
Very recently, some studies employed MALDI-MS to study and distinguish strains of the major variants of concern using mass signatures[83][85]. These studies also demonstrate how such MALDI-MS data can be used to chart viral evolution and to construct mass-based phylogenetic trees, similar to those obtained using conventional gene sequence phylogenetics. The use of MS datasets to track the evolution of SARS-CoV-2 in order to identify such mutations that may limit detection by PCR or those that allow immune escape or reduce the efficacy of existing vaccines and/or therapies have great future potential.
6. Conclusions
Laboratory SARS-CoV-2 testing assays are the primary mitigation measure for the prompt identification and isolation of infected individuals. In fact, the rapid and accurate detection of SARS-CoV-2 infection and the close monitoring of persons in contact with cases are key steps in managing the COVID-19 pandemic, until vaccines can be extensively administered or until antiviral drugs become widely available. In this context, MS-based technologies may represent an alternative resource or may support PCR-based methods as a diagnostic test in the fight against the novel coronavirus. To date, MALDI-MS is routinely used in clinical practice for the rapid, unambiguous, and cost-effective identification of pathogens in biological samples. It is worth noting that several MS-laboratories around the world have been working to deliver guidelines and protocols on sample collection, processing, and data analysis in order to develop rapid and robust methods for the high-throughput screening of SARS-CoV-2 infection. Furthermore, clinical trials using MALDI-MS are currently ongoing for the management of the COVID-19 pandemic. In particular, one of these studies is based on MALDI-TOF MS profiling and combined with ML methods to detect individuals infected with SARS-CoV-2 from saliva samples, while the other one is based on the MALDI-TOF MS analysis of both saliva and blood samples to search for discriminating profiles between COVID-19 and non-COVID-19 individuals. Nevertheless, some limitations still exist for the applications of MALDI-MS technology in routine virus diagnostics. In fact, unlike what happens for bacteria, in the case of biotyping approaches, the major limitations could arise from the relatively low protein content of viruses, the higher molecular weight of viral proteins (>20,000 Da), and a probable carryover of debris from the cell substrate in which the viruses are cultured in vitro.
On the other hand, despite the high costs of the instrument, the high-resolution MALDI MS with proteotyping strategies, based on SARS-CoV-2 signature peptides, has demonstrated excellent performance characteristics especially for the high-accuracy detection of SARS-CoV-2. Considering high accuracy, high throughput, and speed of analysis, the use of more innovative MS-based tools and assays represents an essential and feasible complement to PCR.
MALDI-MS-based genotyping approaches also enable the high-throughput and low-cost detection of SARS-CoV-2. Nevertheless, the level of accuracy of these approaches is not yet comparable to those of proteotyping strategies. In any case, conventional tests such as PCR or immunoassay have become insufficient, also considering the shortages of medium, reagents, collection devices, and consumables that hinder day-to-day laboratory operations and the ability to improve testing capacity. In this scenario, MALDI-MS-based genotyping strategies could represent reliable diagnostic techniques that require alternative consumables. Moreover, recent investigations have demonstrated the ability of these platforms to assess unfolding genomic variation in a timely manner, and have highlighted the potential of diagnostic results to serve as a reliable system for the detection of emergent SARS-CoV-2 variants of concern .
Finally, regarding the proof-of-principle studies, the biomolecular host profiling approaches that assess host response to predict infection are a further intriguing alternative to the current testing methods for the identification of SARS-CoV-2 infected individuals. However, viral load differs among infected individuals, in particular between symptomatic and asymptomatic subjects, which leads to a broad variation in host response. Therefore, these approaches warrant further investigations that would test their threshold for the detection of viral infection and their ability to differentiate types of infectious or non-infectious causes of host modulation, in order to avoid confounding results. Additionally, these strategies also have multiple desirable features for a clinical test during the COVID-19 pandemic, which includes minimal sample preparation, few reagents, and rapid and high-throughput data acquisition.
Arguably, as part of a pandemic preparedness plan, a massive amount of novel scientific knowledge as well as the use of innovative MS-based tools and assays will support the development of diagnostics facilitating a reliable response to emerging infectious diseases.
Certainly, the expanded application of MS-based technologies and related multi-omics strategies could become one of the reference methodologies for other future epidemics/pandemics that may arise.
References
- Coronavirus Update (Live): 92,895,303 Cases and 1,989,457 Deaths from COVID-19 Virus Pandemic—Worldometer . Coronavirus Update (Live). Retrieved 2021-12-10
- Joseph S.M. Peiris; Kwok-Yung Yuen; Albert Osterhaus; Klaus Stöhr; The Severe Acute Respiratory Syndrome. New England Journal of Medicine 2003, 349, 2431-2441, 10.1056/nejmra032498.
- Claude P. Muller; Do asymptomatic carriers of SARS-COV-2 transmit the virus?. The Lancet Regional Health - Europe 2021, 4, 100082-100082, 10.1016/j.lanepe.2021.100082.
- SeyedAhmad SeyedAlinaghi; Pegah Mirzapour; Omid Dadras; Zahra Pashaei; Amirali Karimi; Mehrzad MohsseniPour; Mahdi Soleymanzadeh; Alireza Barzegary; Amir Masoud Afsahi; Farzin Vahedi; et al.Ahmadreza ShamsabadiFarzane BehnezhadSolmaz SaeidiEsmaeil MehraeenShayesteh Jahanfar Characterization of SARS-CoV-2 different variants and related morbidity and mortality: a systematic review. European Journal of Medical Research 2021, 26, 1-20, 10.1186/s40001-021-00524-8.
- Matthias Becker; Alex Dulovic; Daniel Junker; Natalia Ruetalo; Philipp D. Kaiser; Yudi T. Pinilla; Constanze Heinzel; Julia Haering; Bjoern Traenkle; Teresa R. Wagner; et al.Mirjam LayerMartin MehrlaenderValbona MirakajJana HeldHannes PlanatscherKatja Schenke-LaylandGérard KrauseMonika StrengertTamam BakchoulKarina AlthausRolf FendelAndrea KreidenweissMichael KoeppenUlrich RothbauerMichael SchindlerNicole Schneiderhan-Marra Immune response to SARS-CoV-2 variants of concern in vaccinated individuals. Nature Communications 2021, 12, 1-8, 10.1038/s41467-021-23473-6.
- Roshan J. D'cruz; Arthur W. Currier; Valerie B. Sampson; Laboratory Testing Methods for Novel Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-CoV-2). Frontiers in Cell and Developmental Biology 2020, 8, 468, 10.3389/fcell.2020.00468.
- Lauren M. Kucirka; Stephen A. Lauer; Oliver Laeyendecker; Denali Boon; Justin Lessler; Variation in False-Negative Rate of Reverse Transcriptase Polymerase Chain Reaction–Based SARS-CoV-2 Tests by Time Since Exposure. Annals of Internal Medicine 2020, 173, 262-267, 10.7326/m20-1495.
- Ingrid Arevalo-Rodriguez; Diana Buitrago-Garcia; Daniel Simancas-Racines; Paula Zambrano Achig; Rosa Del Campo; Agustin Ciapponi; Omar Sued; Laura Martinez-García; Anne W. Rutjes; Nicola Low; et al.Patrick M. BossuytJose A. Perez-MolinaJavier Zamora False-negative results of initial RT-PCR assays for COVID-19: A systematic review. PLoS ONE 2020, 15, e0242958, 10.1371/journal.pone.0242958.
- Alireza Tahamtan; Abdollah Ardebili; Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Review of Molecular Diagnostics 2020, 20, 453-454, 10.1080/14737159.2020.1757437.
- Adam P Rosebrock; DNA Cross-Reactivity of the CDC-Specified SARS-CoV-2 Specimen Control Leads to Potential for False Negatives and Underreporting of Viral Infection. Clinical Chemistry 2020, 67, 435-437, 10.1093/clinchem/hvaa284.
- Jamil N. Kanji; Nathan Zelyas; Clayton MacDonald; Kanti Pabbaraju; Muhammad Naeem Khan; Abhaya Prasad; Jia Hu; Mathew Diggle; Byron M. Berenger; Graham Tipples; et al. False negative rate of COVID-19 PCR testing: a discordant testing analysis. Virology Journal 2021, 18, 1-6, 10.1186/s12985-021-01489-0.
- Zhenzhou Wan; Yongjuan Zhao; Renfei Lu; Yajuan Dong; Chiyu Zhang; Rapid antigen detection alone may not be sufficient for early diagnosis and/or mass screening of COVID‐19. Journal of Medical Virology 2021, 93, 6462-6464, 10.1002/jmv.27236.
- Rosa Terracciano; Mariaimmacolata Preianò; Annalisa Fregola; Corrado Pelaia; Tiziana Montalcini; Rocco Savino; Mapping the SARS-CoV-2–Host Protein–Protein Interactome by Affinity Purification Mass Spectrometry and Proximity-Dependent Biotin Labeling: A Rational and Straightforward Route to Discover Host-Directed Anti-SARS-CoV-2 Therapeutics. International Journal of Molecular Sciences 2021, 22, 532, 10.3390/ijms22020532.
- Shalini Aggarwal; Arup Acharjee; Amrita Mukherjee; Mark S. Baker; Sanjeeva Srivastava; Role of Multiomics Data to Understand Host–Pathogen Interactions in COVID-19 Pathogenesis. Journal of Proteome Research 2021, 20, 1107-1132, 10.1021/acs.jproteome.0c00771.
- Kevin M. Downard; Bethny Morrissey; Alexander B. Schwahn; Mass spectrometry analysis of the influenza virus. Mass Spectrometry Reviews 2008, 28, 35-49, 10.1002/mas.20194.
- Aleksandra Milewska; Joanna Ner‐Kluza; Agnieszka Dabrowska; Anna Bodzon‐Kulakowska; Krzysztof Pyrc; Piotr Suder; MASS SPECTROMETRY IN VIROLOGICAL SCIENCES. Mass Spectrometry Reviews 2019, 39, 499-522, 10.1002/mas.21617.
- Matthew W. Foster; Geoff Gerhardt; Lynda Robitaille; Pier-Luc Plante; Guy Boivin; Jacques Corbeil; M. Arthur Moseley; Targeted Proteomics of Human Metapneumovirus in Clinical Samples and Viral Cultures. Analytical Chemistry 2015, 87, 10247-10254, 10.1021/acs.analchem.5b01544.
- Wanda I. Santana; Tracie L. Williams; Emily K. Winne; James L. Pirkle; John R. Barr; Quantification of Viral Proteins of the Avian H7 Subtype of Influenza Virus: An Isotope Dilution Mass Spectrometry Method Applicable for Producing more Rapid Vaccines in the Case of an Influenza Pandemic. Analytical Chemistry 2014, 86, 4088-4095, 10.1021/ac4040778.
- Justin H. Griffin; Kevin M. Downard; Mass spectrometry analytical responses to the SARS-CoV2 coronavirus in review. TrAC Trends in Analytical Chemistry 2021, 142, 116328, 10.1016/j.trac.2021.116328.
- Iqbal Mahmud; Timothy J. Garrett; Mass Spectrometry Techniques in Emerging Pathogens Studies: COVID-19 Perspectives. Journal of the American Society for Mass Spectrometry 2020, 31, 2013-2024, 10.1021/jasms.0c00238.
- Imesha W. De Silva; Subhayu Nayek; Vijay Singh; Jay Reddy; John K. Granger; Guido F. Verbeck; Paper spray mass spectrometry utilizing Teslin® substrate for rapid detection of lipid metabolite changes during COVID-19 infection. The Analyst 2020, 145, 5725-5732, 10.1039/d0an01074j.
- Praveen Singh; Rahul Chakraborty; Robin Marwal; V. S. Radhakrishan; Akash Kumar Bhaskar; Himanshu Vashisht; Mahesh S. Dhar; Shalini Pradhan; Gyan Ranjan; Mohamed Imran; et al.Anurag RajUma SharmaPriyanka SinghHemlata LallMeena DuttaParth GargArjun RayDebasis DashSridhar SivasubbuHema GogiaPreeti MadanSandhya KabraSujeet K. SinghAnurag AgrawalPartha RakshitPramod KumarShantanu Sengupta A rapid and sensitive method to detect SARS-CoV-2 virus using targeted-mass spectrometry. Journal of Proteins and Proteomics 2020, 11, 159-165, 10.1007/s42485-020-00044-9.
- Karina Helena Morais Cardozo; Adriana Lebkuchen; Guilherme Gonçalves Okai; Rodrigo Andrade Schuch; Luciana Godoy Viana; Aline Nogueira Olive; Carolina Dos Santos Lazari; Ana Maria Fraga; Celso Francisco Hernandes Granato; Maria Carolina Tostes Pintão; et al.Valdemir Melechco Carvalho Establishing a mass spectrometry-based system for rapid detection of SARS-CoV-2 in large clinical sample cohorts. Nature Communications 2020, 11, 1-13, 10.1038/s41467-020-19925-0.
- Viviana Greco; Cristian Piras; Luisa Pieroni; Maurizio Ronci; Lorenza Putignani; Paola Roncada; Andrea Urbani; Applications of MALDI-TOF mass spectrometry in clinical proteomics. Expert Review of Proteomics 2018, 15, 683-696, 10.1080/14789450.2018.1505510.
- Andrew P. Bowman; Greg T. Blakney; Christopher L. Hendrickson; Shane R. Ellis; Ron M. A. Heeren; Donald F. Smith; Ultra-High Mass Resolving Power, Mass Accuracy, and Dynamic Range MALDI Mass Spectrometry Imaging by 21-T FT-ICR MS. Analytical Chemistry 2020, 92, 3133-3142, 10.1021/acs.analchem.9b04768.
- M. Karas; D. Bachmann; U. Bahr; F. Hillenkamp; Matrix-assisted ultraviolet laser desorption of non-volatile compounds. International Journal of Mass Spectrometry and Ion Processes 1987, 78, 53-68, 10.1016/0168-1176(87)87041-6.
- Craig M. Whitehouse; Robert N. Dreyer; Masamichi. Yamashita; John B. Fenn; Electrospray interface for liquid chromatographs and mass spectrometers. Analytical Chemistry 1985, 57, 675-679, 10.1021/ac00280a023.
- Hillenkamp, F.; Peter-Katalinic, J. The MALDI Process and Method. In MALDI MS: A Practical Guide to Instrumentation, Methods and Application; Wiley-VCH Verlag GmbH & Co. KgaA: Weinheim, Germany, 2007; pp. 1-28.
- Teesha C Baker; Jun Han; Christoph H Borchers; Recent advancements in matrix-assisted laser desorption/ionization mass spectrometry imaging. Current Opinion in Biotechnology 2017, 43, 62-69, 10.1016/j.copbio.2016.09.003.
- Mariia Ivanova; Olena Dyadyk; Dmytro Ivanov; Francesca Clerici; Andrew Smith; Fulvio Magni; Matrix-assisted laser desorption/ionization mass spectrometry imaging to uncover protein alterations associated with the progression of IgA nephropathy. Virchows Archiv 2019, 476, 903-914, 10.1007/s00428-019-02705-7.
- Fei Zhao; Jianzhong Zhang; Xuemei Wang; Liyong Liu; Jie Gong; Zhixiang Zhai; Lihua He; Fanliang Meng; Di Xiao; A multisite SNP genotyping and macrolide susceptibility gene method for Mycoplasma pneumoniae based on MALDI-TOF MS. iScience 2021, 24, 102447, 10.1016/j.isci.2021.102447.
- Clizia Chinello; Marta Cazzaniga; Gabriele De Sio; Andrew James Smith; Erica Gianazza; Angelica Grasso; Francesco Rocco; Stefano Signorini; Marco Grasso; Silvano Bosari; et al.Italo Francesco ZoppisMohammed DaknaYuri E. M. Van Der BurgtGiancarlo MauriFulvio Magni Urinary Signatures of Renal Cell Carcinoma Investigated by Peptidomic Approaches. PLoS ONE 2014, 9, e106684, 10.1371/journal.pone.0106684.
- Rosa Terracciano; Mariaimmacolata Preianò; Giuseppina Maggisano; Corrado Pelaia; Rocco Savino; Hexagonal Mesoporous Silica as a Rapid, Efficient and Versatile Tool for MALDI-TOF MS Sample Preparation in Clinical Peptidomics Analysis: A Pilot Study.. Molecules 2019, 24, 2311, 10.3390/molecules24122311.
- Rosa Terracciano; Mariaimmacolata Preianò; Grazia P. Palladino; Giovanna E. Carpagnano; Maria P. Foschino Barbaro; Girolamo Pelaia; Rocco Savino; Rosario Maselli; Peptidome profiling of induced sputum by mesoporous silica beads and MALDI-TOF MS for non-invasive biomarker discovery of chronic inflammatory lung diseases. PROTEOMICS 2011, 11, 3402-3414, 10.1002/pmic.201000828.
- Mariaimmacolata Preianò; Giuseppina Maggisano; Maria Stella Murfuni; Chiara Villella; Carmela Colica; Annalisa Fregola; Corrado Pelaia; Nicola Lombardo; Girolamo Pelaia; Rocco Savino; et al.Rosa Terracciano Rapid Detection and Identification of Antimicrobial Peptide Fingerprints of Nasal Fluid by Mesoporous Silica Particles and MALDI-TOF/TOF Mass Spectrometry: From the Analytical Approach to the Diagnostic Applicability in Precision Medicine. International Journal of Molecular Sciences 2018, 19, 4005, 10.3390/ijms19124005.
- Nicola Lombardo; Mariaimmacolata Preianò; Giuseppina Maggisano; Maria Stella Murfuni; Luigi Messina; Girolamo Pelaia; Rocco Savino; Rosa Terracciano; A rapid differential display analysis of nasal swab fingerprints to distinguish allergic from non-allergic rhinitis subjects by mesoporous silica particles and MALDI-TOF mass spectrometry. PROTEOMICS 2017, 17, 1600215, 10.1002/pmic.201600215.
- Mariaimmacolata Preianò; Giuseppina Maggisano; Maria Stella Murfuni; Chiara Villella; Corrado Pelaia; Tiziana Montalcini; Nicola Lombardo; Girolamo Pelaia; Rocco Savino; Rosa Terracciano; et al. An Analytical Method for Assessing Optimal Storage Conditions of Gingival Crevicular Fluid and Disclosing a Peptide Biomarker Signature of Gingivitis by MALDI-TOF MS. PROTEOMICS – Clinical Applications 2018, 12, 1800005, 10.1002/prca.201800005.
- Silvia Angeletti; Matrix assisted laser desorption time of flight mass spectrometry (MALDI-TOF MS) in clinical microbiology. Journal of Microbiological Methods 2017, 138, 20-29, 10.1016/j.mimet.2016.09.003.
- Adriana Calderaro; Maria-Cristina Arcangeletti; Isabella Rodighiero; Mirko Buttrini; Chiara Gorrini; Fabio Jose Nascimento Motta; Diego Germini; Maria-Cristina Medici; Carlo Chezzi; Flora De Conto; et al. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry applied to virus identification. Scientific Reports 2014, 4, 6803, 10.1038/srep06803.
- YunJu Cho; Arif Ahmed; Ananna Islam; Sunghwan Kim; Developments in FT-ICR MS instrumentation, ionization techniques, and data interpretation methods for petroleomics. Mass Spectrometry Reviews 2014, 34, 248-263, 10.1002/mas.21438.
- Xiaoshan Sun; Peichun Wu; Chunxia Zhao; Fujian Zheng; Chunxiu Hu; Xin Lu; Guowang Xu; Protein profiling analysis based on matrix-assisted laser desorption/ionization-Fourier transform ion cyclotron resonance mass spectrometry and its application in typing Streptomyces isolates. Talanta 2019, 208, 120439, 10.1016/j.talanta.2019.120439.
- Marialaura Dilillo; R. Ait-Belkacem; C. Esteve; D. Pellegrini; Simone Nicolardi; M. Costa; Eleonora Vannini; Erik L. de Graaf; Matteo Caleo; L. A. McDonnell; et al. Ultra-High Mass Resolution MALDI Imaging Mass Spectrometry of Proteins and Metabolites in a Mouse Model of Glioblastoma. Scientific Reports 2017, 7, 1-11, 10.1038/s41598-017-00703-w.
- Robin Patel; MALDI-TOF MS for the Diagnosis of Infectious Diseases. Clinical Chemistry 2015, 61, 100-111, 10.1373/clinchem.2014.221770.
- Martin Welker; Alex Van Belkum; Victoria Girard; Jean-Philippe Charrier; David Pincus; An update on the routine application of MALDI-TOF MS in clinical microbiology. Expert Review of Proteomics 2019, 16, 695-710, 10.1080/14789450.2019.1645603.
- Malin I. L. Sjöholm; Joakim Dillner; Joyce Carlson; Multiplex Detection of Human Herpesviruses from Archival Specimens by Using Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. Journal of Clinical Microbiology 2008, 46, 540-545, 10.1128/jcm.01565-07.
- Junping Peng; Fan Yang; Zhaohui Xiong; Junhua Guo; Jiang Du; Yongfeng Hu; Qi Jin; Sensitive and rapid detection of viruses associated with hand foot and mouth disease using multiplexed MALDI-TOF analysis. Journal of Clinical Virology 2013, 56, 170-174, 10.1016/j.jcv.2012.10.020.
- Ray Iles; Raminta Zmuidinaite; Jason Iles; George Carnell; Alex Sampson; Jonathan Heeney; Development of a Clinical MALDI-ToF Mass Spectrometry Assay for SARS-CoV-2: Rational Design and Multi-Disciplinary Team Work. Diagnostics 2020, 10, 746, 10.3390/diagnostics10100746.
- Prajkta Chivte; Zane LaCasse; Venkata Devesh R. Seethi; Pratool Bharti; Joshua Bland; Shrihari S. Kadkol; Elizabeth R. Gaillard; MALDI-ToF protein profiling as a potential rapid diagnostic platform for COVID-19. Journal of Mass Spectrometry and Advances in the Clinical Lab 2021, 21, 31-41, 10.1016/j.jmsacl.2021.09.001.
- Petra Wandernoth; Katharina Kriegsmann; Cristina Groh-Mohanu; Martin Daeumer; Peter Gohl; Oliver Harzer; Mark Kriegsmann; Joerg Kriegsmann; Detection of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by Mass Spectrometry. Viruses 2020, 12, 849, 10.3390/v12080849.
- Magda Rybicka; Ewa Miłosz; Krzysztof Bielawski; Superiority of MALDI-TOF Mass Spectrometry over Real-Time PCR for SARS-CoV-2 RNA Detection. Viruses 2021, 13, 730, 10.3390/v13050730.
- Matthew M. Hernandez; Radhika Banu; Paras Shrestha; Armi Patel; Feng Chen; Liyong Cao; Shelcie Fabre; Jessica Tan; Heidi Lopez; Numthip Chiu; et al.Biana ShifrinInessa ZapolskayaVanessa FloresPui Yiu LeeSergio CastañedaJuan David RamírezJeffrey JhangGiuliana OsorioMelissa R. GitmanMichael D. NowakDavid L. ReichCarlos Cordon‐CardoEmilia Mia SordilloAlberto E. Paniz‐Mondolfi RT‐PCR/MALDI‐TOF mass spectrometry‐based detection of SARS‐CoV‐2 in saliva specimens. Journal of Medical Virology 2021, 93, 5481-5486, 10.1002/jmv.27069.
- Nicholas L. Dollman; Justin H. Griffin; Kevin M. Downard; Detection, Mapping, and Proteotyping of SARS-CoV-2 Coronavirus with High Resolution Mass Spectrometry. ACS Infectious Diseases 2020, 6, 3269-3276, 10.1021/acsinfecdis.0c00664.
- Fabiane Manke Nachtigall; Alfredo Pereira; Oleksandra S. Trofymchuk; Leonardo S. Santos; Detection of SARS-CoV-2 in nasal swabs using MALDI-MS. Nature Biotechnology 2020, 38, 1168-1173, 10.1038/s41587-020-0644-7.
- Nam K. Tran; Taylor Howard; Ryan Walsh; John Pepper; Julia Loegering; Brett Phinney; Michelle R. Salemi; Hooman H. Rashidi; Novel application of automated machine learning with MALDI-TOF-MS for rapid high-throughput screening of COVID-19: a proof of concept. Scientific Reports 2021, 11, 1-10, 10.1038/s41598-021-87463-w.
- Meritxell Deulofeu; Esteban García-Cuesta; Eladia María Peña-Méndez; José Elías Conde; Orlando Jiménez-Romero; Enrique Verdú; María Teresa Serrando; Victoria Salvadó; Pere Boadas-Vaello; Detection of SARS-CoV-2 Infection in Human Nasopharyngeal Samples by Combining MALDI-TOF MS and Artificial Intelligence. Frontiers in Medicine 2021, 8, 661358, 10.3389/fmed.2021.661358.
- María Florencia Rocca; Jonathan Cristian Zintgraff; María Elena Dattero; Leonardo Silva Santos; Martín Ledesma; Carlos Vay; Mónica Prieto; Estefanía Benedetti; Martín Avaro; Mara Russo; et al.Fabiane Manke NachtigallElsa Baumeister A combined approach of MALDI-TOF mass spectrometry and multivariate analysis as a potential tool for the detection of SARS-CoV-2 virus in nasopharyngeal swabs. Journal of Virological Methods 2020, 286, 113991-113991, 10.1016/j.jviromet.2020.113991.
- Ling Yan; Jia Yi; Changwu Huang; Jian Zhang; Shuhui Fu; Zhijie Li; Qian Lyu; Yuan Xu; Kun Wang; Huan Yang; et al.Qingwei MaXiaoping CuiLiang QiaoWei SunPu Liao Rapid Detection of COVID-19 Using MALDI-TOF-Based Serum Peptidome Profiling. Analytical Chemistry 2021, 93, 4782-4787, 10.1021/acs.analchem.0c04590.
- Shu-Hua Chen; Christine H. Parker; Timothy R. Croley; Melinda A. McFarland; Identification of Salmonella Taxon-Specific Peptide Markers to the Serovar Level by Mass Spectrometry. Analytical Chemistry 2019, 91, 4388-4395, 10.1021/acs.analchem.8b04843.
- Maria-Theresia Gekenidis; Patrick Studer; Simone Wüthrich; René Brunisholz; David Drissner; Beyond the Matrix-Assisted Laser Desorption Ionization (MALDI) Biotyping Workflow: in Search of Microorganism-Specific Tryptic Peptides Enabling Discrimination of Subspecies. Applied and Environmental Microbiology 2014, 80, 4234-4241, 10.1128/aem.00740-14.
- Kevin M. Downard; Proteotyping for the rapid identification of influenza virus and other biopathogens. Chemical Society Reviews 2013, 42, 8584-8595, 10.1039/c3cs60081e.
- An P. Nguyen; Kevin M. Downard; Proteotyping of the Parainfluenza Virus with High-Resolution Mass Spectrometry. Analytical Chemistry 2013, 85, 1097-1105, 10.1021/ac302962u.
- Reaz Uddin; Kevin M. Downard; Subtyping of hepatitis C virus with high resolution mass spectrometry. Clinical Mass Spectrometry 2017, 4-5, 19-24, 10.1016/j.clinms.2017.08.003.
- Wout Bittremieux; Charlotte Adams; Kris Laukens; Pieter C. Dorrestein; Nuno Bandeira; Open Science Resources for the Mass Spectrometry-Based Analysis of SARS-CoV-2. Journal of Proteome Research 2021, 20, 1464-1475, 10.1021/acs.jproteome.0c00929.
- Jeffrey A SoRelle; Khushbu Patel; Laura Filkins; Jason Y Park; Mass Spectrometry for COVID-19. Clinical Chemistry 2020, 66, 1367-1368, 10.1093/clinchem/hvaa222.
- Mariaimmacolata Preianò; Serena Correnti; Corrado Pelaia; Rocco Savino; Rosa Terracciano; MALDI MS-Based Investigations for SARS-CoV-2 Detection. BioChem 2021, 1, 250-278, 10.3390/biochem1030018.
- Bhavesh D. Kevadiya; Jatin Machhi; Jonathan Herskovitz; Maxim D. Oleynikov; Wilson R. Blomberg; Neha Bajwa; DhruvKumar Soni; Srijanee Das; Mahmudul Hasan; Milankumar Patel; et al.Ahmed M. SenanSanthi GorantlaJoEllyn McMillanBenson EdagwaRobert EisenbergChannabasavaiah B. GurumurthySt Patrick M. ReidChamindie PunyadeeraLinda ChangHoward E. Gendelman Diagnostics for SARS-CoV-2 infections. Nature Materials 2021, 20, 593-605, 10.1038/s41563-020-00906-z.
- Nam K. Tran; Samer Albahra; Tam N. Pham; James H. Holmes IV; David Greenhalgh; Tina L. Palmieri; Jeffery Wajda; Hooman H. Rashidi; Novel application of an automated-machine learning development tool for predicting burn sepsis: proof of concept. Scientific Reports 2020, 10, 1-9, 10.1038/s41598-020-69433-w.
- Nagib Ahsan; R. Shyama Prasad Rao; Rashaun S. Wilson; Ujwal Punyamurtula; Fernanda Salvato; Max Petersen; Mohammad Kabir Ahmed; M. Ruhul Abid; Jacob C. Verburgt; Daisuke Kihara; et al.Zhibo YangLuca FornelliSteven B. FosterBharat Ramratnam Mass spectrometry‐based proteomic platforms for better understanding of SARS‐CoV‐2 induced pathogenesis and potential diagnostic approaches. PROTEOMICS 2021, 21, 2000279, 10.1002/pmic.202000279.
- Paige Haas; Monita Muralidharan; Nevan J. Krogan; Robyn M. Kaake; Ruth Hüttenhain; Proteomic Approaches to Study SARS-CoV-2 Biology and COVID-19 Pathology. Journal of Proteome Research 2021, 20, 1133-1152, 10.1021/acs.jproteome.0c00764.
- Neelja Singhal; Manish Kumar; Pawan Kumar Kanaujia; Jugsharan S. Virdi; MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Frontiers in Microbiology 2015, 6, 791, 10.3389/fmicb.2015.00791.
- Eva Torres-Sangiao; Cristina Leal Rodriguez; Carlos García-Riestra; Application and Perspectives of MALDI–TOF Mass Spectrometry in Clinical Microbiology Laboratories. Microorganisms 2021, 9, 1539, 10.3390/microorganisms9071539.
- Victor M Corman; Olfert Landt; Marco Kaiser; Richard Molenkamp; Adam Meijer; Daniel Kw Chu; Tobias Bleicker; Sebastian Brünink; Julia Schneider; Marie Luisa Schmidt; et al.Daphne MuldersBart L HaagmansBas Van Der VeerSharon Van Den BrinkLisa WijsmanGabriel GoderskiJean-Louis RometteJoanna EllisMaria ZambonMalik PeirisHerman GoossensChantal ReuskenMarion Pg KoopmansChristian Drosten Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045, 10.2807/1560-7917.es.2020.25.3.2000045.
- Daniel K W Chu; Yang Pan; Samuel M S Cheng; Kenrie Pui Yan Hui; Pavithra Krishnan; Yingzhi Liu; Daisy Y M Ng; Carrie K C Wan; Peng Yang; Quanyi Wang; et al.Malik PeirisLeo L M Poon Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clinical Chemistry 2020, 66, 549-555, 10.1093/clinchem/hvaa029.
- Ruiyun Li; Sen Pei; Bin Chen; Yimeng Song; Tao Zhang; Wan Yang; Jeffrey Shaman; Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science 2020, 368, 489-493, 10.1126/science.abb3221.
- Mariaimmacolata Preianò; Daniela Falcone; Giuseppina Maggisano; Tiziana Montalcini; Michele Navarra; Sergio Paduano; Rocco Savino; Rosa Terracciano; Assessment of pre-analytical and analytical variables affecting peptidome profiling of gingival crevicular fluid by MALDI-TOF mass spectrometry. Clinica Chimica Acta 2014, 437, 120-128, 10.1016/j.cca.2014.07.022.
- Mariaimmacolata Preianò; Giuseppina Maggisano; Nicola Lombardo; Tiziana Montalcini; Sergio Paduano; Girolamo Pelaia; Rocco Savino; Rosa Terracciano; Influence of storage conditions on MALDI-TOF MS profiling of gingival crevicular fluid: Implications on the role of S100A8 and S100A9 for clinical and proteomic based diagnostic investigations. PROTEOMICS 2015, 16, 1033-1045, 10.1002/pmic.201500328.
- Lirong Zou; Feng Ruan; Mingxing Huang; Lijun Liang; Huitao Huang; Zhongsi Hong; Jianxiang Yu; Min Kang; Yingchao Song; Jinyu Xia; et al.Qianfang GuoTie SongJianfeng HeHui-Ling YenMalik PeirisJie Wu SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. New England Journal of Medicine 2020, 382, 1177-1179, 10.1056/nejmc2001737.
- Lorenzo Azzi; Giulio Carcano; Francesco Gianfagna; Paolo Grossi; Daniela Dalla Gasperina; Angelo Genoni; Mauro Fasano; Fausto Sessa; Lucia Tettamanti; Francesco Carinci; et al.Vittorio MaurinoAgostino RossiAngelo TagliabueAndreina Baj Saliva is a reliable tool to detect SARS-CoV-2. Journal of Infection 2020, 81, e45-e50, 10.1016/j.jinf.2020.04.005.
- Roman Wölfel; Victor M. Corman; Wolfgang Guggemos; Michael Seilmaier; Sabine Zange; Marcel A. Müller; Daniela Niemeyer; Terry C. Jones; Patrick Vollmar; Camilla Rothe; et al.Michael HoelscherTobias BleickerSebastian BrüninkJulia SchneiderRosina EhmannKatrin ZwirglmaierChristian DrostenClemens Wendtner Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465-469, 10.1038/s41586-020-2196-x.
- Wei Zhang; Rong-Hui Du; Bei Li; Xiao-Shuang Zheng; Xing-Lou Yang; Ben Hu; Yan-Yi Wang; Geng-Fu Xiao; Bing Yan; Zheng-Li Shi; et al.Peng Zhou Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerging Microbes & Infections 2020, 9, 386-389, 10.1080/22221751.2020.1729071.
- Shaun Griffin; Covid-19: Lateral flow tests are better at identifying people with symptoms, finds Cochrane review. BMJ 2021, 372, n823, 10.1136/bmj.n823.
- Congshan Jiang; Xiaowei Li; Changrong Ge; Yuanyuan Ding; Tao Zhang; Shuai Cao; Liesu Meng; Shemin Lu; Molecular detection of SARS-CoV-2 being challenged by virus variation and asymptomatic infection. Journal of Pharmaceutical Analysis 2021, 11, 257-264, 10.1016/j.jpha.2021.03.006.
- Christian Mann; Justin H. Griffin; Kevin M. Downard; Detection and evolution of SARS-CoV-2 coronavirus variants of concern with mass spectrometry. Analytical and Bioanalytical Chemistry 2021, 413, 7241-7249, 10.1007/s00216-021-03649-1.
- William T. Harvey; Alessandro M. Carabelli; Ben Jackson; Ravindra K. Gupta; Emma C. Thomson; Ewan M. Harrison; Catherine Ludden; Richard Reeve; Andrew Rambaut; Sharon J. Peacock; et al.David L. Robertson SARS-CoV-2 variants, spike mutations and immune escape. Nature Reviews Genetics 2021, 19, 409-424, 10.1038/s41579-021-00573-0.
- Matthew M. Hernandez; Radhika Banu; Ana S. Gonzalez‐Reiche; Adriana van de Guchte; Zenab Khan; Paras Shrestha; Liyong Cao; Feng Chen; Huanzhi Shi; Ayman Hanna; et al.Hala AlshammaryShelcie FabreAngela AmoakoAjay OblaBremy AlburquerqueLuz Helena PatiñoJuan David RamírezRobert SebraMelissa R. GitmanMichael D. NowakCarlos Cordon‐CardoTed E. SchutzbankViviana SimonHarm van BakelEmilia Mia SordilloAlberto E. Paniz‐Mondolfi Robust clinical detection of SARS‐CoV‐2 variants by RT‐PCR/MALDI‐TOF multi‐target approach. Journal of Medical Virology 2021, medRxiv 2021, medRxiv 2021, 10.1002/jmv.27510.




