Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Maria Angeles Decena Rodriguez | + 4634 word(s) | 4634 | 2021-12-07 08:42:02 | | | |
| 2 | Maria Angeles Decena Rodriguez | Meta information modification | 4634 | 2021-12-10 15:57:49 | | | | |
| 3 | Vivi Li | Meta information modification | 4634 | 2021-12-13 03:10:58 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Decena Rodriguez, M. Dehydrin Genes in Model Brachypodium Grasses. Encyclopedia. Available online: https://encyclopedia.pub/entry/16990 (accessed on 08 February 2026).
Decena Rodriguez M. Dehydrin Genes in Model Brachypodium Grasses. Encyclopedia. Available at: https://encyclopedia.pub/entry/16990. Accessed February 08, 2026.
Decena Rodriguez, Maria. "Dehydrin Genes in Model Brachypodium Grasses" Encyclopedia, https://encyclopedia.pub/entry/16990 (accessed February 08, 2026).
Decena Rodriguez, M. (2021, December 10). Dehydrin Genes in Model Brachypodium Grasses. In Encyclopedia. https://encyclopedia.pub/entry/16990
Decena Rodriguez, Maria. "Dehydrin Genes in Model Brachypodium Grasses." Encyclopedia. Web. 10 December, 2021.
Copy Citation
Dehydration proteins (dehydrins, DHNs) confer tolerance to water-stress deficit in plants. We performed a comparative genomics and evolutionary study of DHN genes in four model Brachypodium grass species. Ten dehydrins have been describe within Brachypodium species. Due to limited knowledge on dehydrin expression under water deprivation stress in Brachypodium, we also performed a drought-induced gene expression analysis in 32 ecotypes of the genus’ flagship species B. distachyon showing different hydric requirements. Bdhn1 - Bdhn2, Bdhn3 and Bdhn7 genes, orthologs of wheat, barley, rice, sorghum, and maize genes, were more highly expressed in plants under drought conditions.
Bdhn genes
cis-regulatory elements
comparative genomics
dehydrin structure
dehydrin-gene expression
duplicated genes
drought-related traits
drought-tolerant ecotypes
phylogenetics
1. Introduction
Water deprivation is one of the main abiotic stresses that affect plant development and fitness [1][2]. Water deficit stress in plants is mostly caused by low soil water content but also by other abiotic stresses such as salinity and extreme temperature [3][4]—which sometimes interact [2]—affecting the survival of plants in a wide range of environmental conditions [4][5]. Plant species adapted to dry environments have developed mechanisms to protect their cells from water stress deficit. Among the several physiological and genomic regulatory mechanisms triggered by water limitation in plants, there is an almost universal response in the upregulation of dehydrins [1][4][6]. Dehydrins (DHN) belong to group 2 LEA (Late-Embryogenesis-Abundant) proteins [7], and are intrinsically disordered hydrophilic proteins that acquire structure when bound to ligands, such as membranes, acting as chaperones that impede the aggregation or inactivation of other proteins under desiccation to maintain the biological activity of the cell [8][9]. They show a high hydration capacity and can also bind large quantities of cations, retaining water in the drying cells and preventing ionic unbalance and protein denaturation. DHNs are also called RAB proteins because they are usually responsive to abscisic acid [9]. DHNs accumulate in all vegetative tissues under water stress, though with different specificities [4], as well as during seed development [8].
The dehydrin protein family is characterized by a modular sequence domain (Dehydrin) that contains three conserved segments (Y, S, K; [8][10]; Figure 1a). The dehydrin identifier K-segment motif is present in all plant DHNs except for HIRD11 (see Section 2). Perdiguero et al. demonstrated the induction of dehydrin genes lacking the K-segment under drought stress in Pinus [11]. The S-segment is located between the Y and K segments and, when phosphorylated, transfers dehydrins and other nuclear localization signal (NLS) binding proteins from the cytosol to the nucleus [10][12]. The Y-segment motif is located towards the N-terminus of the protein coding region. The Y, S, and K segments are interspersed by other less conserved segments, named interpatter or ϕ-segments that mostly contain small, polar, and charged amino acids [13]. Recent studies have described another conserved F-segment motif in some seed plant DHNs [11][14]. This segment, when present, has a unique copy and is located between the N-terminus of the protein coding region and the S-segment. An additional NLS-segment motif was found in some dehydrin families of maize [12]. Different combinations of conserved motifs have been used to classify the dehydrin domain into major architectures, with five of them (Kn, SKn, KnS, YnKn, YnSKn) being common across angiosperms [10]. The dehydrin domain is occasionally fused with upstream DNAJ-X and DNA-J containing gene domains in some DHN genes [10]. DNA-J is a heat shock protein (Hsp40) that prevents the aggregation of unfolded proteins and functions as a chaperone folding protein when associated with Hsp70 under water-stress [10][15] (Figure 1a). Evolutionary studies of spermatophyte dehydrins identified Kn or SKn as the oldest ancestral architectures whereas those containing the Y segment arose only in the angiosperms [10][16]. Two main architectural changes apparently occurred after the ancestral whole genome duplication events of the angiosperms, the Y3SK2-Y3K1 and Y1SK2-K6 dehydrins [10]. Whereas K6 developed a novel function in cold protection (but see [16] for the important role of Kn dehydrins in drought protection), Y3K1 showed signatures of pseudogenization in some families (e.g., the Brassicaceae) [10]. At the infrageneric level, the dehydrin phylogeny of 11 Oryza species showed the splits of four high-to-weakly supported domain clades and a lack of dehydrin-specific subclades [9].
DHNs have been extensively studied in grasses due to their key role as agents conferring water-stress tolerance in cereal and forage crops [7][9][17][18]. Expression of dehydrins under drought stress has been positively associated with plant biomass and grain yield [19]. Several in vitro studies have demonstrated that DHN expression enhances plant stress tolerance [20][21]. Dehydrins maintain the osmotic balance of cells and their chlorophyll contents, bind metals to scavenge ROS, and bind to DNA and phospholipids [20][22]. Despite these advances, some of the biological functions of DHNs have not been fully established yet [10]. Few dehydrins have been characterized biochemically, like those involved in enzyme and membrane bioprotection in Triticum [23] and Zea [24] and in protection against ROS in Arabidopsis and Brassica [25][26]. Their protective activities have been experimentally demonstrated in some cases using circular dichroism (e.g., Zea mays DHN1 binding to lipid vesicles, [27]; Hordeum vulgare Dhn5 protection of lactate deshydrogenase under drought and cold conditions, [28]).
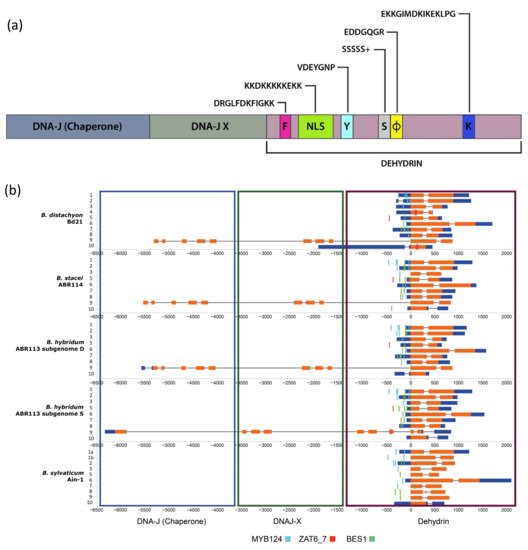
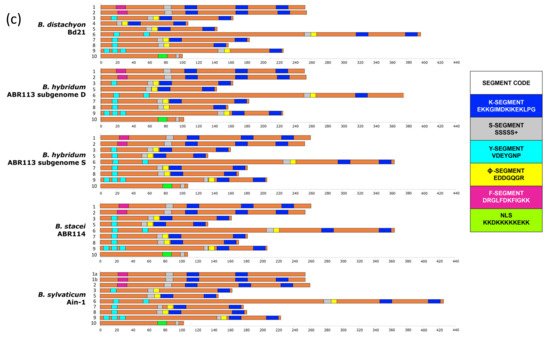
Figure 1. Schematic structure of a typical grass DHN gene showing the positions of the main domains and coding segments (a). Gene structure of Brachypodium distachyon Bd21, B. stacei ABR114, B. hybridum ABR113 (D and S subgenomes), and B. sylvaticum Ain-1 dehydrins showing: (b) the three detected DNA J (purple rectangle), DNA J-X (light green rectangle) and dehydrin (dark green rectangle) domains, and their CDSs (orange bars), introns (grey lines), and 5′ and 3′ UTRs (blue bars); (c) the conserved motifs of their CDSs with their respective K (dark blue), S (grey), Y (light blue), ϕ (yellow), F (pink), and NLS (light green) segments. Brachypodium dehydrin gene codes (Bdhn1–Bdhn10) correspond to those indicated in Table 1. Cis-regulatory elements BES1, MYB124, and ZAT are mapped in the promoters of each gene (see color codes in the chart; the figure could be also visualized in http://zeta.uma.es/public/journal/brachy/DHN_Brachy_4_varieties.html, 29 November 2021).
Table 1. Dehydrin genes found in the studied Brachypodium distachyon, B. stacei, B. hybridum (subgenomes D and S), and B. sylvaticum species. Brachypodium dehydrin gene codes (Bdhn1–Bdhn10) given in this study, the protein structure and their corresponding Panther family and subfamilies gene codes. Phytozome dehydrin gene codes correspond to the respective gene numbers in the reference genome of each species deposited in Phytozome.
| Bdhn | Structure | Panther Family | Panther Subfamily | Phytozome Dehydrin Gene Codes | ||||
|---|---|---|---|---|---|---|---|---|
| B. distachyon (Bd21) |
B. stacei (ABR114) |
B. hybridum D (ABR113) |
B. hybridum S (ABR113) |
B. sylvaticum (Ain1) |
||||
| Bdhn1a | PTH33346 | ERD14 | Bradi5g10860 | Brast09G089800 | Brahy.D05G0138300 | Brahy.S09G0091400 | Brasy9G143400 | |
| Bdhn1b | FSKn | Brasy9G149400 | ||||||
| Bdhn2 | FSKn | Bradi3g51200 | Brast04G110500 | Brahy.D03G0707100 | Brahy.S04G0117200 | Brasy4G117200 | ||
| Bdhn3 | YnSKn | SF23 | Bradi1g37410 | Brast07G152200 | Brahy.D01G0507100 | Brahy.S07G0169500 | Brasy7G144000 | |
| Bdhn4 | YnSKn | SF14 | Bradi4g22280 | |||||
| Bdhn5 | YnSKn | Bradi4g22290 | Brast05G049400 | Brahy.D04G0319400 | Brahy.S05G0054400 | Brasy5G271500 | ||
| Bdhn6 | YnSKn | SF 19 | Bradi4g19525 | Brast05G075400 | Brahy.D04G0277700 | Brahy.S05G0083200 | Brasy4G237500 | |
| Bdhn7 | YnSKn | SF14 | Bradi3g43870 | Brast04G194300 | Brahy.D03G0604200 | Brahy.S04G0208500 | Brasy4G220100 | |
| Bdhn8 | YnSKn | Bradi3g43855 | Brast04G197200 | Brahy.D03G0604000 | Brahy.S04G0208900 | Brasy4G219800 | ||
| Bdhn9 | YnSKn | XERO I | Bradi2g47575 | Brast01G171900 | Brahy.D02G0637300 | Brahy.S01G0182800 | Brasy1G228900 | |
| Bdhn10 | K*NLSL-S | PTHR34941 | HIRD 11 | Bradi1g13330 | Brast02G251900 | Brahy.D01G017200 | Brahy.S02G0268100 | Brasy2G277500 |
Brachypodium is a model system for grasses due to its intermediate evolutionary position between the temperate cereals (Triticeae) and the tropical biofuel (Miscanthus, Paspalum) crops [29]. The three annual species of the genus have been selected as a model complex for polyploidy (diploids B. distachyon and B. stacei and derived allotetraploid B. hybridum; [30]) and one of its perennial species has been selected as a model species for perenniality (diploid B. sylvaticum; [31]). Transgenic plants of its flagship species B. distachyon have been analyzed to identify candidate genes that enhance drought stress tolerance in plants [32][33][34] though none of them specifically addressed dehydrins. Inspection of an early version of the B. distachyon reference genome Bd21 detected 36 LEA2 encoding genes but did not characterize the dehydrin genes [35]. The dehydrin gene content, structure, evolution, and expression in response to drought among species and accessions of Brachypodium has not been investigated yet.
Given the significance of DHNs in water stress response of grasses, we analyzed the members of the dehydrin gene family in the four Brachypodium model species and in 54 B. distachyon ecotypes showing different hydric requirements and drought tolerances using in silico analysis of genome annotations. Due to the lack of studies on dehydrin expression under water deprivation stress in Brachypodium we also performed DHN expression analysis in 32 B. distachyon ecotypes under different drought and watered conditions. The aims of our study were: (i) to identify and characterize the Brachypodium dehydrin genes (Bdhn) and the structure and biochemical properties of the encoded proteins, comparing them with those of the closely related cereal crops (Hordeum, Oryza, Sorghum, Triticum-Aegilops, Zea), and to evaluate the presence of enriched sequence motifs that may have potential regulatory effects (ii) to analyze the syntenic distributions and origins of the Bdhn genes, identify gene duplication events, and test the functionality of paralogs, (iii) to investigate their expression profiles under control vs. dry conditions and compare them to those of closely related cereals, and (iv) to correlate the dehydrin gene expressions with the phenotypic and environmental traits of the plants and test their potential phylogenetic signal.
2. Dehydrin Genes of Brachypodium Species and Outgroup Grasses
Genome searches in Phytozome and Ensembl Plants retrieved 47 dehydrin gene sequences collected from the reference genomes of the four sequenced Brachypodium species [B. distachyon Bd21 2n = 2x = 10, x = 5 (10); B. sylvaticum Ain-1 2n = 2x = 18, x = 9 (10); B. stacei ABR114 2n = 2x = 20, x = 10 (9); B. hybridum ABR113 2n = 4x = 30, x = 5 + 10 (18; 9 from its B. distachyon-type D subgenome and 9 from its B. stacei-type S subgenome)] (Table 1; Figure 1b). A total of 54 orthologous DHN sequences were retrieved from the reference genomes of six outgroup grass species [Aegilops tauschii 2n = 2x = 14, x = 7 (9); Hordeum vulgare 2n = 2x = 14, x = 7 (8); Zea mays 2n = 2x = 20, x = 10 (7); Oryza sativa 2n = 2x = 24, x = 12 (6); Sorghum bicolor 2n = 2x = 20, x = 10 (5); Triticum aestivum 2n = 6x = 42, x = 7 (19); Supplementary Table S1]. A new nomenclature was created for the Brachypodium dehydrin genes (Bdhn1 to Bdhn10; Table 1; Figure 1b,c). In several instances we used the same numbers as those of the orthologous Hordeum vulgare DHN genes (Bdhn 6-7, and Bdhn 9-10) (ENA database: AF043086, AF043092; AF043086 and Genbank database: AY681974) and the orthologous Oryza sativa DHN genes (Bdhn1-2 and Bdhn8) (RAP database: Os02g0669100, Os11g0454300). Bdhn3 was numbered according to prior annotation in Brachypodium (GeneBank: XM_010229280), whereas the remaining Bdhn genes were numbered consecutively as Bdhn4 and Bdhn5. The Bdhn genes were also classified according to Panther protein gene families (PTH33346 and PTH34941) and subfamilies (ERD14, XERO1, SF14, SF19, SF23, HIRD11; Table 1). All Bdhn10 genes found in the studied species of Brachypodium lacked the K-segment (Figure 1c). However, those dehydrins showed an extraordinary sequence identity with typical DHNs, including those with a modified K-segment, like DHN-13 from H. vulgare [36]. The absence of the K-segment has been also reported in one dehydrin of pine species [11] and four dehydrins of rice species (OnDHN6, OrDHN7, OlDHN3, OlDHN6; [9]). These Brachypodium Bdhn10 dehydrins, with architecture K*(NLS)S, belong to HIRD11 proteins and show orthology with DHN genes from H. vulgare, O. sativa, S. bicolor, and Z. mays (Supplementary Table S1).
The Brachypodium dehydrins showed different lengths, ranging from 86.3 (Bdhn10) to 323 amino acid residues (Bdhn6), molecular weights from 9762.90 to 30945.54 kDa, and isoelectric points from 4.41 (Bdhn2) to 7.6 (Bdhn7) (9.4 (Bdhn4) in B. distachyon) (Supplementary Table S2). All dehydrins presented a negative GRAVY value, indicating that they are hydrophilic proteins. All Bdhn genes except Bdhn5 were structurally conserved across the four Brachypodium species (Figure 1b,c). Bdhn4 was only present in B. distachyon, while Bdhn1 was duplicated in B. sylvaticum (Bdhn1a, Bdhn1b) (Table 1; Figure 1b,c). All Bdhn genes but Bdhn9 showed a single dehydrin domain. Bdhn9 encoded a protein of 508 aa with DHN–DNAJ-X–DNA-J domains in all species except in B. sylvaticum, which showed a gene consisting only of the DHN domain (222 aa) (Supplementary Table S2; Figure 1b). Eight gene architectures were found along the Bdhn1–Bdhn10 genes (FSK2, FSK3, SϕK2, YSϕK2, YSϕK, Y3SϕK, Y3SϕK2, NLS-K*S), with YSϕK2 being the most common architecture present in four genes (Bdhn3, Bdhn6, Bdhn7, Bdhn8; Table 1, Figure 1c). Bdhn 3D modeling indicated that the disordered Brachypodium dehydrins lacked tertiary structure whereas their secondary structure consisted of different numbers of α-helices (1–4) and β-sheets in some inferred proteins (Supplementary Figure S1).
Orthology analysis indicates that Bdhn genes were also present in the other surveyed grasses. In total, 6 out of 10 Bdhn genes were found in A. tauschii; Bdhn1-2 copies were also present in H. vulgare and T. aestivum, Bdhn3 in T. aestivum, Bdhn4-5 in H. vulgare, Bdhn7-8 in T. aestivum, Bdhn9 in O. sativa and S. bicolor, and Bdhn10 in H. vulgare, O. sativa, S. bicolor, and Z. mays (Supplementary Table S1). By contrast, five DHN genes of O. sativa (Os01g0702500, Os11g0453900, Os01g0624400, Os01g0225600, Os03g0655400), four of H. vulgare (HORVU6Hr1G083960, HORVU6Hr1G011050, HORVU5Hr1G103460, HORVU3Hr1G089300), and three of Z. mays (Zm00001d017547, Zm00001d043730, Zm00001d013647) had no orthologous sequences in Brachypodium. Pairwise amino acid sequence similarities indicated that Bdhn4 and Bdhn5 were the most similar proteins, followed by Bdhn1 and Bdhn2. Bdhn10 was the most dissimilar dehydrin. Dehydrins with YSϕK2 structure were in general similar to each other both in Brachypodium and in the other grasses.
3. Cis-Regulatory Elements of Bdhn Genes
We performed de novo discovery analysis of cis-regulatory elements (CREs) of Bdhn genes, searching for binding sites of transcription factors (TF) that accumulate around the transcriptional start site and that may control gene expression (Supplementary Figure S2). The analysis consistently identified three clusters, BES1/BZR1, MYB124, and ZAT binding sites (Table 2; Supplementary Figure S2a) that are related with different drought-response signaling pathways. BES1/BZR1 and MYB124 motifs were present in all studied promoters, though MYB124 was predominant in the promoters of Bdhn1 and Bdhn2 and BES1/BZR1in those of Bdhn7 genes and more abundantly in those of the aridic B. stacei and B. hybridum species. By contrast, ZAT was only found in the promoters of Bdhn4, Bdhn5, and Bdhn10 genes in annual species (Table 2; Supplementary Figure S2a).
Table 2. Upstream putative cis-regulatory elements (CREs) found in the promoter region (–500-to-+200 bp) of the Bdhn genes of Brachypodium distachyon, B. stacei, B. hybridum (subgenomes D and S), and B. sylvaticum using Rsat::plants tools and the corresponding reference genome as background. Family identification, motif code (ID), N-cor (normalized correlation), and Sig (significance value) for the highest hit, matches to transcription factor (TF) binding sites, and Bdhn genes with the number of sites found within each species and gene. Species and reference genomes: BD, B. distachyon Bd21; BS, B. stacei ABR114; BHD, B. hybridum subgenome D ABR113; BHS, B. hybridum subgenome S ABR113; BS, B. sylvaticum Ain1. Mapping positions of these cis-regulatory motifs are indicated in Figure 1b.
| Family ID | Motif ID | Ncor | Sig | TF Binding Site | Bdhn Genes (No. Sites Found) |
|---|---|---|---|---|---|
| BRI1-EMS suppressor/brassinazole-resistant | BES1/BZR1 | 0.719 | 4.08 |  |
BD: Bdhn5(1) Bdhn6(1) Bdhn7(3) Bdhn9(1) |
| BHD: Bdhn1(1) Bdhn2(2) Bdhn5(1) Bdhn6(1) Bdhn7(3) Bdhn9(1) | |||||
| BHS: Bdhn1(1) Bdhn2(2) Bdhn5(4) Bdhn6(1) Bdhn7(2) Bdhn8(1) | |||||
| BS: Bdhn1(1) Bdhn2(2) Bdhn5(3) Bdhn6(1) Bdhn7(2) Bdhn8(1) | |||||
| BSY: Bdhn2(1) Bdhn5(1) Bdhn7(1) Bdhn8(2) Bdhn9(1) | |||||
| Myb | MYB124 | 0.598 | 2.18 |  |
BD: Bdhn1(5) Bdhn2(4) Bdhn3(2) Bdhn6(1) Bdhn7(2) Bdhn8(2) |
| BHD: Bdhn1(7) Bdhn2(4) Bdhn3(2) Bdhn6(1) Bdhn7(2) Bdhn8(2) | |||||
| BHS: Bdhn1(7) Bdhn2(4) Bdhn3(2) Bdhn6(1) Bdhn8(2) | |||||
| BS: Bdhn1(8) Bdhn2(4) Bdhn3(2) Bdhn6(1) Bdhn8(2) | |||||
| BSY: Bdhn1a(4) Bdhn1b(4) Bdhn2(4) Bdhn3(2) Bdhn6(1) Bdhn7(2) | |||||
| C2H2 zinc finger | ZAT | 0.738 | 4.91 |  |
BD: Bdhn4(3) Bdhn5(1) Bdhn10(2) |
| BHD: Bdhn5(1) | |||||
| BHS: Bdhn5(1) Bdhn10(1) | |||||
| BS: Bdhn5(2) Bdhn10(1) |
4. The Brachypodium Dehydrin Tree
To explore the evolutionary relationship among dehydrin genes, we constructed a ML Brachypodium dehydrin gene tree from 47 Bdhn protein coding regions present in the four studied Brachypodium species and in six outgroup grasses (Figure 2). All internal branches but three and all the dehydrin-type clades showed strong to relatively high bootstrap support values (BS ≥ 70%). The most divergent split separated the duplicated Bdhn1-Bdhn2 (ERD14) clade from the rest, followed by the isolated Bdhn10 (HIRD11) clade. Within the remaining group of YnSKn dehydrin structural genes, there was a divergence of the Bdhn9 (XEROI) clade, followed by subsequent divergences of the YnSKn Bdhn4-Bdhn5, Bdhn6, Bdhn3, and Bdhn7/Bdhn8 clades (Figure 2). All Bdhn clades were monophyletic except the paraphyletic Bdhn9 and Bdhn10 clades, which included orthologous sequences from closely related outgroups. Intra-clade branches were overall well-supported except for the poorly informative Bdhn10 clade.
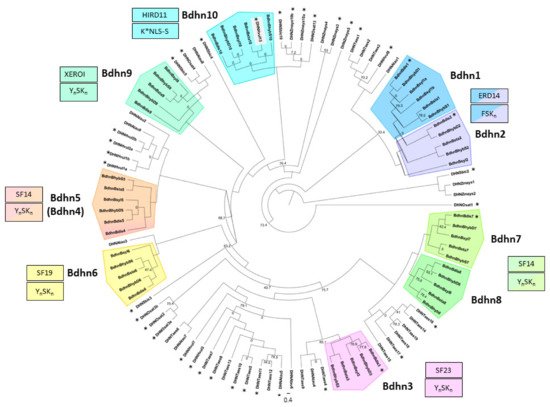
Figure 2. Maximum likelihood Brachypodium Bdhn tree. Unrooted IQtree cladogram showing the relationships among the dehydrin Bdhn gene clades and orthologous grass sequences (Atau: Aegilops tauchii; Hvul: Hordeum vulgare; Osat: Oryza sativa; Sbic: Sorghum bicolor; Taes: Triticum aestivum; Zmays: Zea mays) and among Brachypodium species and genomes within each clade. Bdhn clades are identified by colors, Bdhn codes, gene architecture, and Panther subfamily codes (see Table 1). Duplicated Bdhn genes form sister clades or fall within the same clade. Ultrafast bootstrap support (<80%) is shown on branches; the remaining branches are fully supported. Asterisks indicate dehydrin genes differentially expressed under drought vs. control conditions (see text and Supplementary Table S10). Scale bar: number of mutations per site.
5. Chromosome Distributions and Selection Analysis of Duplicated Bdhn Genes
We analyzed the physical distributions of Bdhn genes on the chromosomes of the studied species to detect the potential occurrence of tandem or segmental duplication. We also performed selection tests on the coding regions of the Bdhn genes to explore the potential loss of selective constrains on them. Dehydrin genes were distributed among the five chromosomes of B. distachyon Bd21 and the D subgenome of B. hybridum ABR113 (Bd), in 6 out of 10 chromosomes of B. stacei ABR114 and the S subgenome of B. hybridum ABR113 (Bs), and in 6 out of 9 chromosomes of B. sylvaticum Ain-1 (Bsy) (Figure 3, Supplementary Table S3). We detected four tandem duplications and two segmental duplications of Bdhn genes. In B. distachyon and B. hybridum D subgenome our analysis detected tandem duplications of Bdhn7-Bdhn8 in Bd3 and of Bdhn4-Bdhn5 in Bd4 (only B. distachyon), and a segmental duplication of Bdhn1-Bdhn2 in Bd3 and Bd5 (Figure 3, Supplementary Table S3). B. stacei and B. hybridum S subgenome and B. sylvaticum showed a tandem duplication of Bdhn7-Bdhn8 in Bs4 and Bsy4, and a segmental duplication of Bdhn1-Bdhn2 in Bs4 and Bs9 and in Bsy4 and Bsy9, respectively. In addition, B. sylvaticum showed a tandem duplication of Bdhn1a and Bdhn1b in Bsy9 not found in the other Brachypodium species studied (Figure 3, Supplementary Table S3). Overall, all the selection tests performed with branch-sites models (aBSREL (adaptive Branch-Site Random Effects Likelihood) and BUSTED (Branch-Site Unrestricted Statistical Test for Episodic Diversification) for internal branches only and for leaf branches only) failed to detect evidence of positive selection in all the Bdhn genes and species all (p-values >0.05) except for two significant cases. aBSREL tests for internal branches only detected evidence of positive selection in Bdhn6 and Bdhn10 (one branch each) where ω2 rate class values were >1 but for very low percentages of tree branches (Bdhn6: p = 0.04, ω1 = 0.140 (97%), ω2 = 53.9 (2.6%); Bdhn10: p = 0.02, ω1 = 0.0 (99%), ω2 = 100,000 (1.1%); Supplementary Materials S1). Whereas aBSREL tests for positive selection modeling both site-level and branch-level ω heterogeneity, BUSTED performs a gene-wide (and not site-specific) test for positive selection. Similarly, selection tests performed at sites [MEME (Mixed Effects Model of Evolution)] did not detect evidence of positive selection at any site across the sequences of the Bdhn genes except for two significant or marginally significant positions in genes Bdhn1-Bdhn2 (site 139, p = 0.04; site 163, p = 0.02) and two in Bdhn6 (site 266, p = 0.04; site 412, p = 0.05) (Supplementary Materials S1). The greater power of MEME indicates that selection acting at individual sites is considerably more widespread than constant models would suggest. It also suggests that natural selection is predominantly episodic, with transient periods of adaptive evolution masked by the prevalence of purifying or neutral selection on other branches.
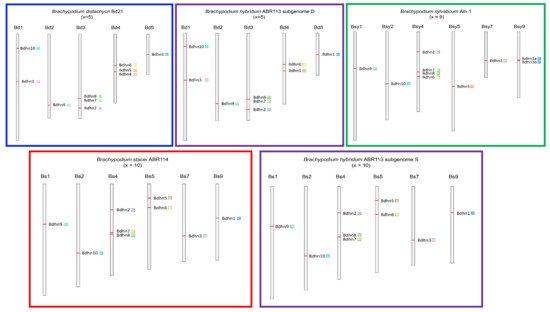
Figure 3. Chromosomal location of Bdhn genes in the studied Brachypodium species. B. distachyon (blue rectangle), B. stacei (red), B. hybridum subgenomes S and D (purple), and B. sylvaticum (green). Bdhn genes are mapped on the chromosome with their respective color flags (see Figure 2).
6. Dehydrin Gene Clusters, Phylogenetics, and Climate Niche Variation in B. distachyon
The genomes of 54 B. distachyon ecotypes distributed across the Mediterranean region (Supplementary Table S4) were annotated for the 10 Bdhn gene clusters for comparative genomics, evolutionary, and phylogenetic signal of drought-related traits and climate niche analyses. Most ecotypes (74.07%) contained all 10 Bdhn genes (Supplementary Figure S3) and were used for downstream analyses. Independent ML trees obtained for each separate Bdhn gene, based on exon and intron sequences, showed differently resolved topologies (Supplementary Figure S4). Six of those 10 gene trees (Bdhn1, Bdhn2, Bdhn3, Bdhn6, Bdhn7, Bdhn8) recovered a congruent topology for some B. distachyon groups. A ML tree constructed from their concatenated sequences produced a single combined B. distachyon Bdhn tree (Figure 4a) showing a resolution similar to those observed previously in the B. distachyon nuclear-SNPs [37] (Figure 4b) and plastome [38] (Figure 4c) trees. The Bdhn tree revealed a relatively well-supported EDF+ clade (74% BS) and the successive but weakly supported divergences of the S+ and T+ lineages, with a clade of T+ lineages (Bd18-1, Bd21-3, BdTR5i) resolved as sister to the EDF+ clade (Figure 4a). Topological congruence Kishino-Hasekawa (KH), Shimodaira-Hasekawa (SH), and Shimodaira Approximately Unbiased (AU) tests performed between the B. distachyon Bdhn tree and the nuclear-SNP and plastome trees indicated that the topology of the Bdhn tree did not significantly differ (p < 0.001) from the topologies of the two compared trees (Supplementary Table S5), indicating that all three data sets recover congruent evolutionary histories for the divergences of the main B. distachyon lineages. However, the Bdhn tree visually resembled more the plastome tree than the nuclear tree (Figure 4).
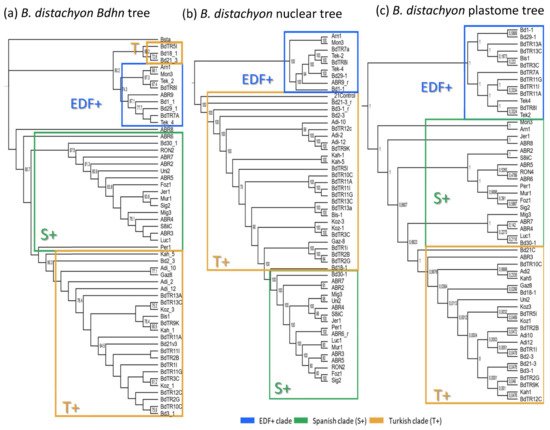
Figure 4. (a) Maximum likelihood B. distachyon Bdhn tree. Consensus tree constructed for 54 B. distachyon ecotypes from concatenated exon and intron aligned sequences of six dehydrin genes (Bdhn1, Bdhn2, Bdhn3, Bdhn6, Bdhn7, and Bdhn8). The EDF+ (extremely delayed flowering time, blue) clade, T+ (Turkish and East Mediterranean, orange), and S+ (Spain and West Mediterranean, green) lineages correspond to those indicated in [37][38]. Bootstrap support is indicated on branches. Accession codes correspond to those indicated in Supplementary Table S4. (b) B. distachyon nuclear species tree of [37] based on nuclear genome-wide 3.9 million SNPs. (c) B. distachyon plastome tree of [38] based on full plastome sequences.
Climate niches were constructed for the studied B. distachyon ecotypes to explore if they present differences in climate niche parameters that could be related to distinct responses to drought. The optimal climate niche of each B. distachyon ecotype was inferred from PCA of 19 climate variables (Supplementary Table S6 and Figure S5). The main PC1 axis (51.8% of variance) values allowed us to classify the B. distachyon ecotypes into cold (>2.5; ABR2, ABR3, ABR4, ABR5, ABR6, RON2), warm (<2.5; Bd2-3, Bd21, Bd3-1, Bis1, Kah1, Kah5, Koz1, Koz3), and mesic (−2.5 to 2.5, remaining) climate class ecotypes.
7. Differential Expression of Bdhn Genes in Brachypodium distachyon Ecotypes under Drought and Temperature Stress Conditions
We made use of data from an extensive transcriptome study of 32 B. distachyon ecotypes [39], which enabled us to explore intraspecific variation in how Bdhn genes are expressed in response to drought in mature plants. Development and tissue specific expression analysis of dehydrin genes was performed in 32 out of the 54 genomically sequenced ecotypes of B. distachyon (Supplementary Table S4). Only 4 out of the 10 identified Bdhn genes were expressed in mature leaves of all 32 studied B. distachyon ecotypes: Bdhn1a (Bradi5g10860.1), Bdhn2 (Bradi3g51200.1), Bdhn3 (Bradi1g37410.1), Bdhn7 (Bradi3g43870.1); Supplementary Table S7. These annotated dehydrins showed significant differential expression (DE) levels between the watered (W) and dry (D) conditions, independently of temperature conditions in both separate CD-CW-HD-HW and averaged D vs. W comparative tests (Supplementary Table S8 and Figure S6a,b). By contrast, the dehydrin expressions did not show significant differences between cool (C) and hot (H) conditions under drought treatment, and only Bdhn3 and Bdhn7 showed significant differences in CW vs. HW conditions, though none of them did in averaged C vs. H comparative tests (Supplementary Figure S6a,c).
The four dehydrins showed significantly increased expression levels in drought conditions in most accessions (Wilcoxon tests, Supplementary Table S8; Tukey tests, Figure 5, Supplementary Figure S7). The DE levels were also significantly different among ecotypes, especially within the dry treatment, being highest in warm ecotypes Koz3 (Bdhn1a, Bdhn3, Bdhn7) and Adi10 (Bdhn2) and lowest in cold ecotypes ABR2 (Bdhn1) and ABR4 (Bdhn2, Bdhn3, Bdhn7) (Figure 5, Supplementary Figure S7). On average drought increased dehydrin expression by about 5.74% in Bdhn1a, 39% in Bdhn2, 67.8% in Bdhn3, and 97.8% in Bdhn7 compared to well-watered plants (Supplementary Figure S6b). Overexpression of dehydrins caused by drought stress was significantly correlated between all Bdhn gene pairs (Supplementary Table S9 and Figure S8).
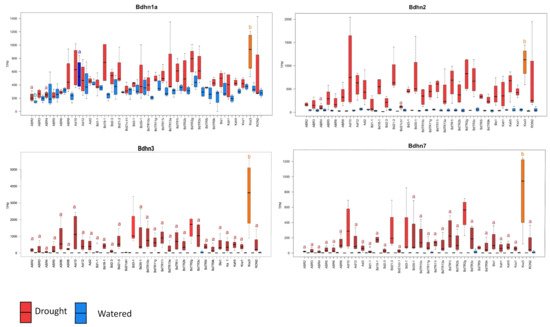
Figure 5. Differentially expressed Bdhn1a, Bdhn2, Bdhn3, and Bdhn7 dehydrin genes (transcript per million, TPM) in 32 ecotypes of B. distachyon under drought (D, red) vs. watered (W, blue) conditions. Different letters in the boxplots indicate significant group differences (Tukey HSD tests) (see also Supplementary Figure S7).
A B. distachyon–T. aestivum DE comparative analysis found that the drought-induced Bdhn1, Bdhn2, Bdhn3, and Bdhn7 genes belong to the same ortholog groups as 15 out of the 16 differentially expressed wheat dehydrin (DHN) genes (Supplementary Table S10) under natural field drought stress [40] or greenhouse imposed drought stress [6][41]. In wheat there is a physical clustering of several dehydrin genes in two gene clusters located in the 5L and 6L groups of wheat chromosomes (Supplementary Table S10; Supplementary Figure S9). The 6L cluster contains 25 dehydrins and includes nine DHN3 genes and three DHN4 genes (all orthologs to Bdhn3), whereas the 5L cluster has 13 DHN38 genes of which six are orthologs to Bdhn7. In addition, the DHN11 genes, located in another portion of 6L chromosomes are orthologs to the Bdhn1 and Bdhn2 genes. We observed that orthologs from B. distachyon and T. aestivum tended to show a similar pattern of expression response to soil drying. Specifically, the duplicated Bdhn1 and Bdhn2 genes and the DHN11(A1) genes, the Bdhn7 gene and the duplicated DHN38 (B1, B2) genes, and the Bdhn3 gene and the duplicated DHN3 (A1, A6, B6, D1, D4, D6, D8, D9) genes were all upregulated in drought treatments (Supplementary Table S10 and Figure S9; [40]).
8. Effects of Drought on Dehydrin Gene Expression and Drought-Response Phenotypic Traits
The potential effect of drought on dehydrin expression levels and on correlated changes of phenotypic and physiological drought-response traits of the plants was evaluated in 32 B. distachyon ecotypes. The 12 drought-response phenotypic traits studied (leaf_rwc: relative water content in leaf; leaf_wc: water content in leaf; lma: leaf mass per area; pro: leaf proline content; abvgrd: above ground biomass; blwgrd: below ground biomass; ttlmass: total plant mass; rmr: root mass ratio; delta13c: carbon isotope, a proxy for lifetime integrated WUE; leafc: leaf carbon content; leafn: leaf nitrogen content; cn: leaf carbon/nitrogen ratio) also showed significant different values in dry vs. watered conditions across ecotypes (Supplementary Table S11). On average, drought significantly decreased the values of six traits (17.14% in abvgrd, 34.78% in ttlmass, 4% in leaf_rwc, 36.5% in leaf_wc, 12.5% in leafn, 2.8% in WUE) and significantly increased those of five traits (33% in pro, 5.5% in rmr, 2.96% in leafc, 5.71% in lma, 21.4% in cn) compared to watered conditions, but did not significantly affect the blwgrd trait (Supplementary Figure S10) [2].
Drought-induced effects caused significant positive and negative correlations between the averaged expressed values of the four Bdhn genes and changes in most phenotypic trait values (Supplementary Table S12 and Figure S11). Regression models for independent correlations between the Bdhn1a, Bdhn2, Bdhn3, and Bdhn7 expressions and the changes in the 12 phenotypic traits showed significant positive correlations for most dehydrin Bdhn genes with pro, blwgrd, rmr, WUE, leafc, and cn, negative correlations with leaf_rwc, leaf_wc, and leafn, and non-significant correlations with ttlmass and abvgrd (Supplementary Figure S11).
9. Phylogenetic Signal of Dehydrin Expression, Phenotypic Trait Changes and Climate Variation in the Brachypodium distachyon Bdhn Tree
The potential phylogenetic signal of dehydrin expression, phenotypic trait changes, and climate variation was evaluated on both the B. distachyon nuclear species tree [37] and the B. distachyon dehydrin Bdhn tree. None of the dehydrin gene expression values under W or D conditions and few phenotypic and climate traits had significant K or lambda values on the B. distachyon nuclear species tree (Supplementary Table S13a and Figure S12). By contrast, Bdhn2W, Bdhn7W, and Bdhn3D expression values, phenotypic change of rmrW, leafnW, cnW, abwgrdW, leafcD, leafnD, and cnD traits’ values and climate niche PCA1 values carried phylogenetic signal (or marginal phylogenetic signal for leaf_rwcD and abvgrdD values) when tested on the dehydrin Bdhn tree (Supplementary Table S13b, Figure 6).
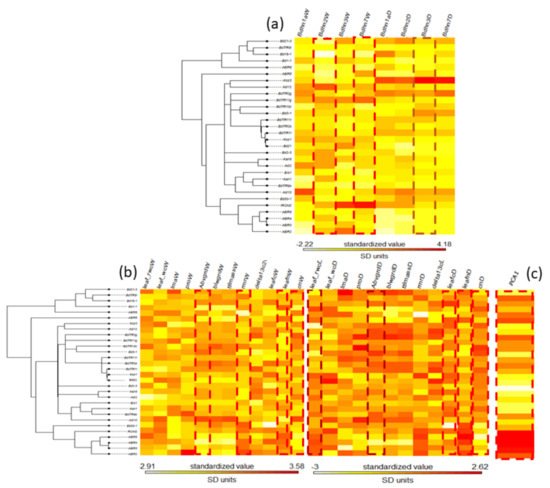
Figure 6. Maximum likelihood B. distachyon Bdhn tree cladogram showing the relationships of 30 ecotypes. Phyloheatmaps of normalized values for different sets of variables: (a) dehydrin (Bdhn1, Bdhn2, Bdhn3, Bdhn7) gene expression values under watered (W) and drought (D) conditions; (b) drought-response phenotypic traits (leaf_rwc: relative water content in leaf; leaf_wc: water content in leaf; lma: leaf mass per area; pro: leaf proline content; abvgrd: above ground biomass; blwgrd: below ground biomass; ttlmass: total plant mass; rmr: root mass ratio; delta13c: carbon isotope, a proxy for lifetime integrated WUE; leafc: leaf carbon content; leafn: leaf nitrogen content; cn: leaf carbon/nitrogen ratio) values under watered (W) and drought (D) conditions; (c) climate niche PC1 values. Traits showing significant phylogenetic signal are highlighted with dotted lines (see Supplementary Table S13b). Watered (W): soil irrigated to field capacity every second day; Drought (D): soil water content reduced by ~5% each day (during the 10 days experiment).
References
- Hossain, M.A.; Wani, S.H.; Bhattacharjee, S.; Burritt, D.J.; Tran, L.-S.P. Drought Stress Tolerance in Plants; Springer: Cham, Switzerland, 2016; Volume 2.
- Des Marais, D.L.; Lasky, J.R.; Verslues, P.E.; Chang, T.Z.; Juenger, T.E. Interactive effects of water limitation and elevated temperature on the physiology, development and fitness of diverse accessions of Brachypodium distachyon. New Phytol. 2017, 214, 132–144.
- Mahajan, S.; Tuteja, N. Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 2005, 444, 139–158.
- Hanin, M.; Brini, F.; Ebel, C.; Toda, Y.; Takeda, S.; Masmoudi, K. Plant dehydrins and stress tolerance: Versatile proteins for complex mechanisms. Plant Signal. Behav. 2011, 6, 1503–1509.
- Tommasini, L.; Svensson, J.T.; Rodriguez, E.M.; Wahid, A.; Malatrasi, M.; Kato, K.; Wanamaker, S.; Resnik, J.; Close, T.J. Dehydrin gene expression provides an indicator of low temperature and drought stress: Transcriptome-based analysis of Barley (Hordeum vulgare L.). Funct. Integr. Genom. 2008, 8, 387–405.
- Reddy, S.K.; Liu, S.; Rudd, J.C.; Xue, Q.; Payton, P.; Finlayson, S.A.; Mahan, J.; Akhunova, A.; Holalu, S.V.; Lu, N. Physiology and transcriptomics of water-deficit stress responses in wheat cultivars TAM 111 and TAM 112. J. Plant Physiol. 2014, 171, 1289–1298.
- Wang, Y.; Xu, H.; Zhu, H.; Tao, Y.; Zhang, G.; Zhang, L.; Zhang, C.; Zhang, Z.; Ma, Z. Classification and expression diversification of wheat dehydrin genes. Plant Sci. 2014, 214, 113–120.
- Graether, S.P.; Boddington, K.F. Disorder and function: A review of the dehydrin protein family. Front. Plant Sci. 2014, 5, 1–12.
- Verma, G.; Dhar, Y.V.; Srivastava, D.; Kidwai, M.; Chauhan, P.S.; Bag, S.K.; Asif, M.H.; Chakrabarty, D. Genome-wide analysis of rice dehydrin gene family: Its evolutionary conservedness and expression pattern in response to PEG induced dehydration stress. PLoS ONE 2017, 12, e0176399.
- Riley, A.C.; Ashlock, D.A.; Graether, S.P. Evolution of the modular, disordered stress proteins known as dehydrins. PLoS ONE 2019, 14, e0211813.
- Perdiguero, P.; Collad, C.; Soto, Á. Novel dehydrins lacking complete K-segments in Pinaceae. The exception rather than the rule. Front. Plant Sci. 2014, 5, 682.
- Goday, A.; Jensen, A.B.; Culiáñez-Macià, F.A.; Albà, M.M.; Figueras, M.; Serratosa, J.; Torrent, M.; Pagès, M. The maize abscisic acid-responsive protein Rab17 is located in the nucleus and interacts with nuclear localization signals. Plant Cell 1994, 6, 351–360.
- Rosales, R.; Romero, I.; Escribano, M.I.; Merodio, C.; Sanchez-Ballesta, M.T. The crucial role of Φ- and K-segments in the in vitro functionality of Vitis vinifera dehydrin DHN1a. Phytochemistry 2014, 108, 17–25.
- Strimbeck, G.R. Hiding in plain sight: The F segment and other conserved features of seed plant SKn dehydrins. Planta 2017, 245, 1061–1066.
- Hennessy, F.; Nicoll, W.S.; Zimmermann, R.; Cheetham, M.E.; Blatch, G.L. Not all J domains are created equal: Implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 2005, 14, 1697–1709.
- Malik, A.A.; Veltri, M.; Boddington, K.F.; Singh, K.K.; Graether, S.P. Genome analysis of conserved dehydrin motifs in vascular plants. Front. Plant Sci. 2017, 8, 1–18.
- Kosová, K.; Vítámvás, P.; Prášil, I.T. Wheat and barley dehydrins under cold, drought, and salinity—What can LEA-II proteins tell us about plant stress response? Front. Plant Sci. 2014, 5, 343.
- Suprunova, T.; Krugman, T.; Fahima, T.; Chen, G.; Shams, I.; Korol, A.; Nevo, E. Differential expression of dehydrin genes in wild barley, Hordeum spontaneum, associated with resistance to water deficit. Plant Cell Environ. 2004, 27, 1297–1308.
- Karami, A.; Shahbazi, M.; Niknam, V.; Shobbar, Z.S.; Tafreshi, R.S.; Abedini, R.; Mabood, H.E. Expression analysis of dehydrin multigene family across tolerant and susceptible barley (Hordeum vulgare L.) genotypes in response to terminal drought stress. Acta Physiol. Plant 2013, 35, 2289–2297.
- Yu, Z.; Wang, X.; Zhang, L. Structural and functional dynamics of dehydrins: A plant protector protein under abiotic stress. Int. J. Mol. Sci. 2018, 19, 3420.
- Lv, A.; Su, L.; Liu, X.; Xing, Q.; Huang, B.; An, Y.; Zhou, P. Characterization of Dehydrin protein, CdDHN4-L and CdDHN4-S, and their differential protective roles against abiotic stress in vitro. BMC Plant Biol. 2018, 18, 299.
- Liu, Y.; Song, Q.; Li, D.; Yang, X.; Li, D. Multifunctional roles of plant dehydrins in response to environmental stresses. Front. Plant Sci. 2017, 8, 1018.
- Drira, M.; Saibi, W.; Brini, F.; Gargouri, A.; Masmoudi, K.; Hanin, M. The K-segments of the wheat dehydrin DHN-5 are essential for the protection of lactate dehydrogenase and β-glucosidase activities in vitro. Mol. Biotechnol. 2013, 54, 643–650.
- Xing, X.; Liu, Y.; Kong, X.; Liu, Y.; Li, D. Overexpression of a maize dehydrin gene, ZmDHN2b, in tobacco enhances tolerance to low temperature. Plant Growth Regul. 2011, 65, 109–118.
- Xu, J.; Zhang, Y.X.; Wei, W.; Han, L.; Guan, Z.Q.; Wang, Z.; Chai, T.Y. BjDHNs confer heavy-metal tolerance in plants. Mol. Biotechnol. 2008, 38, 91–98.
- Hara, M.; Kondo, M.; Kato, T. A KS-type dehydrin and its related domains reduce Cu-promoted radical generation and the histidine residues contribute to the radical-reducing activities. J. Exp. Bot. 2013, 64, 1615–1624.
- Koag, M.C.; Fenton, R.D.; Wilkens, S.; Close, T.J. The binding of maize DHN1 to lipid vesicles. Gain of structure and lipid specificity. Plant Physiol. 2003, 131, 309–316.
- Hughes, S.L.; Schart, V.; Malcolmson, J.; Hogarth, K.A.; Martynowicz, D.M.; Tralman-Baker, E.; Patel, S.N.; Graether, S.P. The importance of size and disorder in the cryoprotective effects of dehydrins. Plant Physiol. 2013, 163, 1376–1386.
- Scholthof, K.-B.G.; Irigoyen, S.; Catalán, P.; Mandadi, K.K. Brachypodium: A monocot grass model system for plant biology. Plant Cell 2018, 30, 1673–1694.
- Gordon, S.P.; Contreras-Moreira, B.; Levy, J.J.; Djamei, A.; Czedik-Eysenberg, A.; Tartaglio, V.S.; Session, A.; Martin, J.; Cartwright, A.; Katz, A.; et al. Gradual polyploid genome evolution revealed by pan-genomic analysis of Brachypodium hybridum and its diploid progenitors. Nat. Commun. 2020, 11, 3670.
- Steinwand, M.A.; Young, H.A.; Bragg, J.N.; Tobias, C.M.; Vogel, J.P. Brachypodium sylvaticum, a Model for Perennial Grasses: Transformation and Inbred Line Development. PLoS ONE 2013, 8, e75180.
- Lee, K.; Kang, H. Emerging roles of RNA-binding proteins in plant growth, development, and stress responses. Mol. Cells 2016, 39, 179–185.
- Ryu, H.; Cho, Y.G. Plant hormones in salt stress tolerance. J. Plant Biol. 2015, 58, 147–155.
- Yoon, J.S.; Kim, J.Y.; Lee, M.B.; Seo, Y.W. Over-expression of the Brachypodium ASR gene, BdASR4, enhances drought tolerance in Brachypodium distachyon. Plant Cell Rep. 2019, 38, 1109–1125.
- Filiz, E.; Ozyigit, I.I.; Tombuloglu, H.; Koc, I. In silico comparative analysis of LEA (Late Embryogenesis Abundant) proteins in Brachypodium distachyon L. Plant Omi. J. 2013, 6, 433–440.
- Rodríguez, E.M.; Svensson, J.T.; Malatrasi, M.; Choi, D.W.; Close, T.J. Barley Dhn13 encodes a KS-type dehydrin with constitutive and stress responsive expression. Theor. Appl. Genet. 2005, 110, 852–858.
- Gordon, S.P.; Contreras-Moreira, B.; Woods, D.P.; Des Marais, D.L.; Burgess, D.; Shu, S.; Stritt, C.; Roulin, A.; Schackwitz, W.; Tyler, L.; et al. Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nat. Commun. 2017, 8, 2184.
- Sancho, R.; Cantalapiedra, C.P.; López-Alvarez, D.; Gordon, S.P.; Vogel, J.P.; Catalán, P.; Contreras-Moreira, B. Comparative plastome genomics and phylogenomics of Brachypodium: Flowering time signatures, introgression and recombination in recently diverged ecotypes. New Phytol. 2018, 218, 1631–1644.
- Sancho, R.; Catalán, P.; Contreras-Moreira, B.; Juenger, T.E.; Des Marais, D.L. Patterns of gene co-expression under water-deficit treatments and pan-genome occupancy in Brachypodium distachyon. bioRxiv 2021.
- Gálvez, S.; Mérida-García, R.; Camino, C.; Borrill, P.; Abrouk, M.; Ramírez-González, R.H.; Biyiklioglu, S.; Amil-Ruiz, F.; Dorado, G.; Budak, H.; et al. Hotspots in the genomic architecture of field drought responses in wheat as breeding targets. Funct. Integr. Genom. 2019, 19, 295–309.
- Chu, C.; Wang, S.; Paetzold, L.; Wang, Z.; Hui, K.; Rudd, J.C.; Xue, Q.; Ibrahim, A.M.H.; Metz, R.; Johnson, C.D.; et al. RNA-seq analysis reveals different drought tolerance mechanisms in two broadly adapted wheat cultivars ‘TAM 111′ and ‘TAM 112’. Sci. Rep. 2021, 11, 4301.
More
Information
Subjects:
Agronomy; Evolutionary Biology; Genetics & Heredity
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Entry Collection:
Environmental Sciences
Revisions:
3 times
(View History)
Update Date:
13 Dec 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




