Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jean marie François | + 2907 word(s) | 2907 | 2021-11-26 07:14:04 | | | |
| 2 | Dean Liu | Meta information modification | 2907 | 2021-12-13 01:47:08 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
François, J.M. FLO11p. Encyclopedia. Available online: https://encyclopedia.pub/entry/16987 (accessed on 07 February 2026).
François JM. FLO11p. Encyclopedia. Available at: https://encyclopedia.pub/entry/16987. Accessed February 07, 2026.
François, Jean Marie. "FLO11p" Encyclopedia, https://encyclopedia.pub/entry/16987 (accessed February 07, 2026).
François, J.M. (2021, December 10). FLO11p. In Encyclopedia. https://encyclopedia.pub/entry/16987
François, Jean Marie. "FLO11p." Encyclopedia. Web. 10 December, 2021.
Copy Citation
Flo11p may be taken as a model for understanding cell–cell and cell–surface adhesion mechanisms that are exploited by pathogenic yeasts to adhere to abiotic surfaces such as catheters and gain access to the internal organs of patients or to serve as a reservoir of drug-resistant infectious cells in the form of biofilms.
cell wall
flocculins
FLO11
adaptive plasticity
1. Primary Sequence Analysis Distinguishes Flo11p from the Other Yeast Flocculins
The S. cerevisiae flocculins belong to a large family of fungal glycosylphosphoinositide-linked cell wall proteins (GPI-CWPs). As illustrated in Figure 1, these flocculins, which are encoded by 5 genes, exhibit a modular architecture with an N-terminal domain (A-domain) flanked by a secretory signal sequence, a large middle region (B-domain) containing serine/threonine-rich repeats, and a C-terminal part at which a remnant of GPI anchor establishes the cross-link between an amino acid of this C-domain and a non-reducing end of β-1,6-glucan, which involves a transamidation and transglycosylation reactions, respectively [1]. The representation of the primary structure of the flocculins by a hydrophobic-cluster analysis (HCA) [2] already points to differences between Flo11p and the other flocculins. First, on a global sequence, the similarity between Flo1p and Flo11p is only 37% whereas it is >90% between Flo1p, Flo5p, and Flo9p [3]. Second, the A-domain that spans about 200–230 amino acids at the N-terminus of the flocculin is very different in sequence and structure between Flo11p and the other flocculins. For Flo1 to Flo10p, this domain encompasses a PA14 lectin domain, which is widely distributed in the Bacteria, Archaea, and Eukarya domain, and harbors a unique Ca2+-binding motif DcisD. It also contains a so called Flo subdomain that is exposed at the cell surface and bears the carbohydrate binding site and [4][5]. This type of A-domain fully accounts for the dominant Ca2+-dependent flocs formation. For further details on the structure of the P14-Flo domain, the reader should consult the recent review on adhesins in yeast [6].
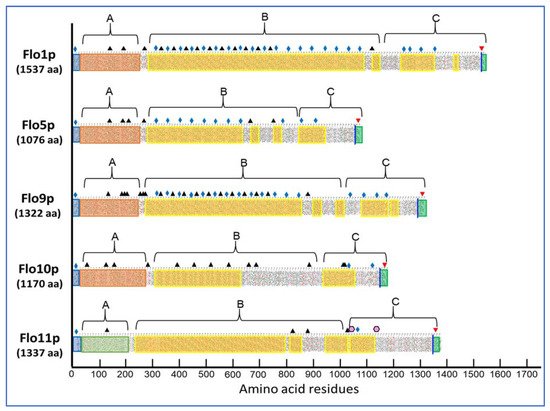
Figure 1. Primary structure analysis of the flocculins encoded by the gene family of the Saccharomyces cerevisiae strain S288c as represented by a hydrophobic-cluster analysis (HCA).
In contrast, the structural analysis of the A-domain of Flo11p has revealed a fibronectin type III-like adhesion domain that does not have any mannose-binding sites [7]. Interestingly, this type of domain is found only in the ascomycetal orders of Saccharomycetales, and in the Flo11p of Komagataella pastoris (formerly known as Pichia pastoris [8]) although sharing only 32% homology with ScFlo11p [9]. Interestingly, this A-domain is present in up to three times in adhesins of the human pathogenic Candida lusitaniae and the wood-boring beetle associated fungus, Spasthasphora passalidarum [7].
A third significant difference between Flo11p and the other flocculins is found at the large middle region (B-domain) of these proteins. Albeit the primary sequence in this B-domain consists of tandem repeats, this analysis eventually splits the flocculins into two categories. The first one that comprises Flo1p, Flo5p, and Flo9p is characterized by a large number of short sequences of 5 to 7 amino acids especially rich in β-branched amino acids Val, Ile, and Thr with a β-aggregation potential > 30% as predicted by TANGO predictor software (http://tango.crg.es/) (accessed on 15 October 2021) More than 50% of these β-aggregation prone sequences have the consensus T(V/I)IVI and they are all present in the 45-residue repeats of these flocculins. It is considered that high β-aggregation potential sequences provide amyloid fiber properties to these proteins [10]. Ramsook et al. [11] indeed showed that a synthetic peptide containing a Flo1p TANGO positive sequence forms amyloid fibers in vitro. The second category encompasses Flo10p and Flo11p, which exhibit less β-aggregation complexity within the B-domain. However, Flo11p possesses at its C-terminus two β-aggregation sequences that nicely match the amyloid-core sequence VVSTTV or VTTAVT, which is supported by the finding that this protein is also able to aggregate into amyloid fibers in vitro [11][12] and in vivo ([13] see below).
2. Relationship between Sequence/Structure and Role in the Physiological Function of Flo11p
2.1. Strain Phenotypes Are Shaped by the Plasticity of the FLO11-Encoded Protein
The gene encoding Flo11p was originally isolated and characterized independently by two research groups from a Saccharomyces cerevisiae var. diastaticus [14][15], which is a yeast able to grow on starch due to the production of a glucoamylase encoded by any one of the members of the STA genes family [16]. Two seminal properties of this flocculin were already disclosed in these works, which were the requirement of Flo11p for invasive and pseudohyphal growth under carbon or nitrogen limitation and its involvement in flocculation. Subsequent work revealed other phenotypes, including the formation of velum, which is a biofilm of yeast cells floating on the surface of a wine barrel [17][18][19], and mats, which correspond to a floral-like biofilm that expands over a wet, semi-solid surface by sliding motility [20]. Adhesion of yeast cells on solid surfaces such as plastic or polystyrene was reported as a property elicited by Flo11p [21]. Additionally, Flo11p was shown to be implicated in colony morphologies exhibited by different yeast strains growing on solid media in the presence of various carbon sources [22]. More recently, we showed that Flo11 molecules could cluster together to form adhesion nanodomains on the cell surface, a property that is dependent on a threshold number of amyloid-β-aggregation prone sequences in the Flo11p ([23]; discussed below). This wide variety of phenotypes raises the question of whether they can be expressed collectively in a single yeast strain and which domain(s) of Flo11p is/are responsible for these phenotypes. Works from the Hyman [24] and Verstrepen teams [25] have in part provided some answers to the first question. They both used the laboratory strain S288c in which FLO genes are not expressed because of a nonsense mutation in the major transcriptional activator encoded by FLO8 [26]. While one group integrated individual FLO genes under the strong TEF promoter, the other expressed FLO11 in a high copy plasmid under the galactose inducible GAL1 promoter. These works led to two major findings regarding differences between Flo11p and the other flocculins. On the one hand, only Flo11p has the ability to trigger strong invasive growth in a semi-solid environment, and thus it is essential in mats formation [27]. However, this property only occurs if the FLO8 gene is functional, suggesting that while the absence of FLO11 prevents the invasion process, the latter requires other factors under the control of FLO8. On the other hand, FLO11 is unable to induce a flocculation phenotype, whether FLO8 is active or not, although agglutination of 10–30 cells can be observed under an optical microscope in a strain that expresses only this gene, indicating that Flo11p promotes cell–cell interactions but not to the extent that it induces aggregation of thousands of cells leading to flocculation. Since this result was at variance to Flo11p-mediated flocculation of the S. cerevisiae var. diastaticus strain [28], it was argued that strain-specific difference in the Flo11p phenotype may result from significant sequence differences in the FLO11 alleles, rather than quantitative differences in FLO11 expression [29]. This assertion was further supported by the fact that laboratory strain S288c is unable to exhibit a flor phenotype or to produce nanodomains on its cell surface even upon dramatic overexpression of FLO11 [23]. In conclusion, these results highlight the importance of the sequence/structure of the Flo11 protein in the expression of these phenotypes.
2.2. Role of A, B, and C-Domains in the Physiological Function of Flo11p
As exposed in Figure 1, the Flo11p presents a modular architecture in three domains or regions whose structure–function began to emerge from recent works [7][23][29][30][31]. The N-terminal domain of Flo11p (A-domain also referred as N-Flo11p) of the S288c strain from aa 22 to 207 was purified and its crystal structure solved at 0.89 Å resolution. This N-Flo11p was shown to be composed of three subdomains: a hydrophobic apical region, a β-sandwich of fibronectin type III domain, and a neck domain [7]. The N-Flo11p can be O-glycosylated but not N-glycosylated; although, there is a predicted N-glycosylated site (Figure 1) and it did not harbor any carbohydrate binding site. The relevant property of the N-Flo11p is to show a homophilic Flo1p interaction. This interaction was nicely demonstrated in vitro using surface plasmon resonance analysis [32] and in vivo using mutants that express Flo11p defective of the N-terminus by single-cell force spectroscopy experiments [31]. Cells of these mutants do not interact each other anymore and they also have lost the invasive growth phenotype [7]. More remarkably, this N-Flo11p confers kin discrimination at the species and subspecies level, accounting for social behavior of yeast by allowing aggregation between single cells expressing the same Flo11p and excluding those that do not express Flo11p or a different alleles of FLO11 as well as cells from different yeast species that express a paralog of ScFLO11 [31]. This homophilic interaction depends on evolutionary conserved aromatic amino acids residues, which form two bands that are present at the apical and the neck region of the N-Flo11p [7]. How different N-Flo11p variants can discriminate between homotypic (kin) and heterotypic (non-kin) interactions is still unclear. Nonetheless, this property makes FLO11 a member of the green beard genes, which confers cooperation between cells that carry the same allele [33]. These exquisite structural details of the N-terminal of Flo11 and their role in adhesion still leave unexplained why Flo11p from S. cerevisiae var. diastaticus causes flocculation, mediates adherence to plastic, but does not elicit agar invasion [28], even though this protein shows homotypic interaction [12]. Barua et al. [29] compared primary sequence of Flo11p from this strain with that of S288c and Σ1278b. Apart a 15-amino acid insertion in the N-terminal of the Flo11p of Σ1278b, which is otherwise exclusively found in the ‘sake lineage’ of the S. cerevisiae strain [34] and confers a higher adhesion force between cells [31], no other difference could be noticed between the N-terminus of these three Flo11p. This could suggest that another genetic component whose function is dependent on the presence of Flo11p is implicated in the phenotypes shown by S. cerevisiae var. diastaticus.
The Flo11p harbors at its carboxyl terminal a typical GPI-attachment site “GAANIKVLGNFMWLLLALPVVF” that is composed of the ω-site (G) at position 1346 followed by a hydrophobic-like sequence termed pro-peptide (aa 1347 to 1376) [35]. This attachment signal is cleaved off and replaced by a preformed GPI-anchor in the endoplasmic reticulum, which enables trafficking of the modified protein via the classical secretory pathway to end up at the plasma membrane as GPI-PMPs [9]. A still unresolved issue is how some of these GPI-PMPs are sorted into the cell walls to be covalently linked to β-1,6-glucan, which involves a transglycosylation mediated by a Dfg5 enzyme [36][37]. Based on an in silico analysis of 51 protein sequences with putative GPI attachment sites, Caro et al. [35] proposed that the presence of two basic amino acids upstream of the ω site at the C-terminus of these GPI-anchored proteins would in most cases dictate retention at the membrane, whereas GPI-PMPs that do not have these motifs, such as Flo11p, are transferred to the cell wall. Another model is that of Vogt et al. [37] who propose that it is the ethanolamine phosphate (EtN-P) modifications on the central GPI glycan that partly dictate the transfer of GPI-PMPs to the cell wall, with the enzyme Dfg5 acting as a decoder of the presence of the EtN-P modification on these GPI-glycans. Whatever the model, the destination of a GPI-anchored protein to the membrane or to the cell wall is not absolute. Accordingly, it was reported that Flo11 protein defective in the C-terminus can be secreted into the culture medium [12], whereas our recent immunofluorescence experiments showed that such a Flo11p variant was still localized at the cell surface.
These results indicate that part of Flo11p could be weakly retained in the cell wall by non-covalent bonding mainly by hydrogen bonding and S-S bridges [38]. Hot SDS-β-mercaptoethanol treatment of the yeast cell wall should be tested to validate this hypothesis [39]. Moreover, the finding that Flo11p can shed from the cells may also agree with the fact that not all Flo11 proteins are covalently attached to the β-1,6-glucan Moreover, this shedding involves a cleavage within the N-terminus of Flo11p by the protease Kex2p [40]. Nonetheless, the major consequence of losing the C-domain of Flo11p is that a yeast strain expressing this variant is no longer able to grow invasively in a semi-solid medium and to form mats, and same effects that were found in a kex2 mutant [23][40]. The loss of these phenotypes can be explained by the inability of the cells to be retained on the agar plate after washing since this Flo11p variant is no longer retained covalently on the cell wall.
Although there are less data on the structure and function of the central part of the S. cerevisiae Flo11p protein, referred as the B-domain (Figure 1), than that of the N- and C-terminus, several studies attest to its strategic importance in the function of Flo11p. The B-domain contains several tandem repeated Ser/Thr sequences predicted to be O- and N-glycosylated, but they are comparatively less numerous than Flo1p, which could explain the higher hydrophobicity character of the latter [41]. This B-domain is also critically important for the velum phenotype of the flor strains of S. cerevisiae as reported by Fidalgo et al. [17]. These authors found that the Flo11p of the flor strain 133d had twice as many tandem repeats as its counterpart in the laboratory strain S288c and that this higher number of repeats was accompanied by increased cell hydrophobicity, suggesting that this gain in hydrophobicity was critical to the floatability property of this strain. The same argument of cell surface hydrophobicity to elicit this buoyancy property was proposed by Zara et al. [19]. In addition, these authors showed a good correlation between length of the B-domain and potency of making this type of biofilm. This conclusion was completed by Fidalgo et al. [42] who indicated that in addition to the length variation in repeats, changes in the order and/or proportion of the different repeats in Flo11p may be another factor contributing to the buoyant biofilm as well as to other phenotypes including adherence to plastic and invasion in agar. Our recent work carried out with a Flo11p from an industrial strain L69 led to a similar conclusion [23]. Finally, the N-glycosylation status of the B-domain may be another source of phenotypic variation brought about by Flo11p. Using a conditional pmi40-101 mutant, which is compromised in the early stage of glycosylation due to the loss of phosphomannose isomerase activity at the restrictive temperature, Meem and Cullen [43] showed a significant reduction of invasive growth, mat formation, and pseudohyphal development. It would be worth to verify whether velum formation is also abolished in flor strain defective in this process.
2.3. Dual Role of the Amyloid-Forming Sequence in Flo11p-Dependent Cell–Cell Interactions
Fungal adhesins, including the S. cerevisiae flocculins have several β-aggregation-prone sequences that consist mostly of aliphatic β-branched amino acids, which have a high propensity to form β-aggregates. This β-aggregation of proteins increases their local concentration and consequently increases the avidity of binding [44][45]. Physically, the formation of these clusters can be elicited by shear force, such as vortex mixing, laminar flow, or stretching in atomic force microscopy (AFM) [46][47][48]. Moreover, they can be visualized as high avidity adhesins patches termed nanodomains by AFM [48][49]. In the pathogenic yeast, Candida albicans, this phenomenon has received a lot of attention because cell surface adhesins control essential processes of adhesion, colonization, and biofilm formation on host tissues and indwelling medical catheters [50]. Work from Lipke and colleagues have shown that these amyloid-core sequences are needed for clustering adhesin molecules in cis on the cell surface, but they also mediate cell–cell interaction in trans through these cross-β bonds [51][52]. The Flo11p from the laboratory strains S288c and Σ1278b has two typical amyloid core sequences (VVSTTV/VTTAVT) at the boundary between the middle region and the C-terminal domain (see Figure 1), which may likely explain that a soluble version of this protein can assemble into amyloid fibers in vitro [11][12]. In spite of in vitro data and in vivo experiments showing increased cell–cell aggregation upon hydrodynamic shear that can be antagonized by amyloidophilic perturbants [11][13][47], the finding of amyloid-dependent formation of nanodomains and their role in cell–cell adhesion have not been directly demonstrated in the yeast S. cerevisiae, until we discovered the formation of abundant patches on the cell surface of an industrial wine yeast L69 strain under the contact of an AFM bare tip [53]. We demonstrated that these patches corresponded to adhesion nanodomains formed by the clustering of Flo11p, which exhibited nanomechanical properties similar to those formed by nanodomains of Candida albicans adhesins [49]. The formation of nanodomains on the cell surface of the L69 strain and not on that of the laboratory strain S288c even after overexpression of its endogenous Flo11p was explained by a duplication of a short 100 amino acids sequence near the C-terminal of Flo11p of the industrial strain, which provided two additional amyloid-core sequences (VVSTTV). This data argued that a threshold number of these short β-aggregation prone sequences is necessary for effective nanodomains production under shear force [23]. Moreover, this work provided evidence that amyloid-core sequences contribute to trans-interactions between Flo11p of opposing cells, and hence supported the model of Lipke and colleagues for a dual role of amyloid β-sheet interactions; that is, in the formation of clusters of Flo11p on the cell surface (cis-interaction) and in homophilic bonding between Flo11p of opposing cells (trans-interactions) [52][54].
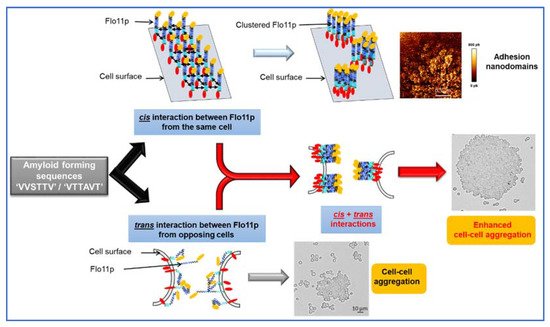
Figure 2. Model by which amyloid-β-aggregation sequences can contribute to nanodomains by cis-interaction of Flo11p molecules and cell–cell aggregation by trans-interaction of Flo11p of opposing cells.
References
- Orlean, P. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 2012, 192, 775–818.
- Lemesle-Varloot, L.; Henrissat, B.; Gaboriaud, C.; Bissery, V.; Morgat, A.; Mornon, J.P. Hydrophobic cluster analysis: Procedures to derive structural and functional information from 2-d-representation of protein sequences. Biochimie 1990, 72, 555–574.
- Dranginis, A.M.; Rauceo, J.M.; Coronado, J.E.; Lipke, P.N. A biochemical guide to yeast adhesins: Glycoproteins for social and antisocial occasions. Microbiol. Mol. Biol. Rev. 2007, 71, 282–294.
- Goossens, K.V.; Stassen, C.; Stals, I.; Donohue, D.S.; Devreese, B.; De Greve, H.; Willaert, R.G. The n-terminal domain of the flo1 flocculation protein from saccharomyces cerevisiae binds specifically to mannose carbohydrates. Eukaryot. Cell 2011, 10, 110–117.
- Veelders, M.; Bruckner, S.; Ott, D.; Unverzagt, C.; Mosch, H.U.; Essen, L.O. Structural basis of flocculin-mediated social behavior in yeast. Proc. Natl. Acad. Sci. USA 2010, 107, 22511–22516.
- Willaert, R.G. Adhesins of yeasts: Protein structure and interactions. J. Fungi 2018, 4, 119.
- Kraushaar, T.; Bruckner, S.; Veelders, M.; Rhinow, D.; Schreiner, F.; Birke, R.; Pagenstecher, A.; Mosch, H.U.; Essen, L.O. Interactions by the fungal flo11 adhesin depend on a fibronectin type iii-like adhesin domain girdled by aromatic bands. Structure 2015, 23, 1005–1017.
- Kurtzman, C.P. Description of komagataella phaffii sp. Nov. And the transfer of Pichia pseudopastoris to the methylotrophic yeast genus komagataella. Int. J. Syst Evol. Microbiol. 2005, 55, 973–976.
- Essen, L.O.; Vogt, M.S.; Mosch, H.U. Diversity of GPI-anchored fungal adhesins. Biol. Chem. 2020, 401, 1389–1405.
- Fernandez-Escamilla, A.M.; Rousseau, F.; Schymkowitz, J.; Serrano, L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004, 22, 1302–1306.
- Ramsook, C.B.; Tan, C.; Garcia, M.C.; Fung, R.; Soybelman, G.; Henry, R.; Litewka, A.; O’Meally, S.; Otoo, H.N.; Khalaf, R.A.; et al. Yeast cell adhesion molecules have functional amyloid-forming sequences. Eukaryot. Cell 2010, 9, 393–404.
- Douglas, L.M.; Li, L.; Yang, Y.; Dranginis, A.M. Expression and characterization of the flocculin flo11/muc1, a Saccharomyces cerevisiae mannoprotein with homotypic properties of adhesion. Eukaryot. Cell 2007, 6, 2214–2221.
- Chan, C.X.; El-Kirat-Chatel, S.; Joseph, I.G.; Jackson, D.N.; Ramsook, C.B.; Dufrene, Y.F.; Lipke, P.N. Force sensitivity in saccharomyces cerevisiae flocculins. mSphere 2016, 1, e00128-16.
- Lo, W.S.; Dranginis, A.M. Flo11, a yeast gene related to the sta genes, encodes a novel cell surface flocculin. J. Bacteriol. 1996, 178, 7144–7151.
- Lambrechts, M.G.; Bauer, F.F.; Marmur, J.; Pretorius, I.S. Muc1, a mucin-like protein that is regulated by mss10, is critical for pseudohyphal differentiation in yeast. Proc. Natl. Acad. Sci. USA 1996, 93, 8419–8424.
- Pretorius, I.S.; Lambrechts, M.G.; Marmur, J. The glucoamylase multigene family in Saccharomyces cerevisiae var. Diastaticus: An overview. Crit. Rev. Biochem. Mol. Biol. 1991, 26, 53–76.
- Fidalgo, M.; Barrales, R.R.; Ibeas, J.I.; Jimenez, J. Adaptive evolution by mutations in the flo11 gene. Proc. Natl. Acad. Sci. USA 2006, 103, 11228–11233.
- Ishigami, M.; Nakagawa, Y.; Hayakawa, M.; Iimura, Y. Flo11 is the primary factor in flor formation caused by cell surface hydrophobicity in wild-type flor yeast. Biosci. Biotechnol. Biochem. 2006, 70, 660–666.
- Zara, S.; Bakalinsky, A.T.; Zara, G.; Pirino, G.; Demontis, M.A.; Budroni, M. Flo11-based model for air-liquid interfacial biofilm formation by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2005, 71, 2934–2939.
- Reynolds, T.B.; Fink, G.R. Bakers’ yeast, a model for fungal biofilm formation. Science 2001, 291, 878–881.
- Guo, B.; Styles, C.A.; Feng, Q.; Fink, G.R. A saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 2000, 97, 12158–12163.
- Voordeckers, K.; De Maeyer, D.; van der Zande, E.; Vinces, M.D.; Meert, W.; Cloots, L.; Ryan, O.; Marchal, K.; Verstrepen, K.J. Identification of a complex genetic network underlying Saccharomyces cerevisiae colony morphology. Mol. Microbiol. 2012, 86, 225–239.
- Bouyx, C.; Schiavone, M.; Teste, M.A.; Dague, E.; Sieczkowski, N.; Julien, A.; Francois, J.M. The dual role of amyloid-beta-sheet sequences in the cell surface properties of flo11-encoded flocculins in Saccharomyces cerevisiae. elife 2021, 10, e68592.
- Purevdorj-Gage, B.; Orr, M.E.; Stoodley, P.; Sheehan, K.B.; Hyman, L.E. The role of flo11 in Saccharomyces cerevisiae biofilm development in a laboratory based flow-cell system. FEMS Yeast Res. 2007, 7, 372–379.
- Van Mulders, S.E.; Christianen, E.; Saerens, S.M.; Daenen, L.; Verbelen, P.J.; Willaert, R.; Verstrepen, K.J.; Delvaux, F.R. Phenotypic diversity of flo protein family-mediated adhesion in Saccharomyces cerevisiae. FEMS Yeast Res. 2009, 9, 178–190.
- Liu, H.; Styles, C.A.; Fink, G.R. Saccharomyces cerevisiae s288c has a mutation in flo8, a gene required for filamentous growth. Genetics 1996, 144, 967–978.
- Reynolds, T.B.; Jansen, A.; Peng, X.; Fink, G.R. Mat formation in Saccharomyces cerevisiae requires nutrient and ph gradients. Eukaryot. Cell 2008, 7, 122–130.
- Bayly, J.C.; Douglas, L.M.; Pretorius, I.S.; Bauer, F.F.; Dranginis, A.M. Characteristics of flo11-dependent flocculation in saccharomyces cerevisiae. FEMS Yeast Res. 2005, 5, 1151–1156.
- Barua, S.; Li, L.; Lipke, P.N.; Dranginis, A.M. Molecular basis for strain variation in the saccharomyces cerevisiae adhesin flo11p. mSphere 2016, 1, e00129-16.
- Goossens, A.; Forment, J.; Serrano, R. Involvement of nst1p/ynl091w and msl1p, a u2b” splicing factor, in Saccharomyces cerevisiae salt tolerance. Yeast 2002, 19, 193–202.
- Bruckner, S.; Schubert, R.; Kraushaar, T.; Hartmann, R.; Hoffmann, D.; Jelli, E.; Drescher, K.; Muller, D.J.; Oliver Essen, L.; Mosch, H.U. Kin discrimination in social yeast is mediated by cell surface receptors of the flo11 adhesin family. elife 2020, 9, e55587.
- Goossens, K.V.; Willaert, R.G. The n-terminal domain of the flo11 protein from Saccharomyces cerevisiae is an adhesin without mannose-binding activity. FEMS Yeast Res. 2012, 12, 78–87.
- West, S.A.; Griffin, A.S.; Gardner, A.; Diggle, S.P. Social evolution theory for microorganisms. Nat. Rev. Microbiol. 2006, 4, 597–607.
- Liti, G.; Carter, D.M.; Moses, A.M.; Warringer, J.; Parts, L.; James, S.A.; Davey, R.P.; Roberts, I.N.; Burt, A.; Koufopanou, V.; et al. Population genomics of domestic and wild yeasts. Nature 2009, 458, 337–341.
- Caro, L.H.; Tettelin, H.; Vossen, J.H.; Ram, A.F.; van den, E.H.; Klis, F.M. In silicio identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 1997, 13, 1477–1489.
- Kitagaki, H.; Wu, H.; Shimoi, H.; Ito, K. Two homologous genes, dcw1 (ykl046c) and dfg5, are essential for cell growth and encode glycosylphosphatidylinositol (GPI)-anchored membrane proteins required for cell wall biogenesis in Saccharomyces cerevisiae. Mol. Microbiol. 2002, 46, 1011–1022.
- Vogt, M.S.; Schmitz, G.F.; Varon Silva, D.; Mosch, H.U.; Essen, L.O. Structural base for the transfer of GPI-anchored glycoproteins into fungal cell walls. Proc. Natl. Acad. Sci. USA 2020, 117, 22061–22067.
- Klis, F.M. Review: Cell wall assembly in yeast. Yeast 1994, 10, 851–869.
- Frieman, M.B.; Cormack, B.P. Multiple sequence signals determine the distribution of glycosylphosphatidylinositol proteins between the plasma membrane and cell wall in Saccharomyces cerevisiae. Microbiology 2004, 150, 3105–3114.
- Karunanithi, S.; Vadaie, N.; Chavel, C.A.; Birkaya, B.; Joshi, J.; Grell, L.; Cullen, P.J. Shedding of the mucin-like flocculin flo11p reveals a new aspect of fungal adhesion regulation. Curr. Biol. 2010, 20, 1389–1395.
- Verbelen, P.J.; Dekoninck, T.M.; Saerens, S.M.; Van Mulders, S.E.; Thevelein, J.M.; Delvaux, F.R. Impact of pitching rate on yeast fermentation performance and beer flavour. Appl. Microbiol. Biotechnol. 2009, 82, 155–167.
- Fidalgo, M.; Barrales, R.R.; Jimenez, J. Coding repeat instability in the flo11 gene of saccharomyces yeasts. Yeast 2008, 25, 879–889.
- Meem, M.H.; Cullen, P.J. The impact of protein glycosylation on flo11-dependent adherence in saccharomyces cerevisiae. FEMS Yeast Res. 2012, 12, 809–818.
- Lipke, P.N.; Garcia, M.C.; Alsteens, D.; Ramsook, C.B.; Klotz, S.A.; Dufrene, Y.F. Strengthening relationships: Amyloids create adhesion nanodomains in yeasts. Trends Microbiol. 2012, 20, 59–65.
- Lipke, P.N.; Ovalle, R. Cell wall architecture in yeast: New structure and new challenges. J. Bacteriol. 1998, 180, 3735–3740.
- Garcia, M.C.; Lee, J.T.; Ramsook, C.B.; Alsteens, D.; Dufrene, Y.F.; Lipke, P.N. A role for amyloid in cell aggregation and biofilm formation. PLoS ONE 2011, 6, e17632.
- Chan, C.X.; Lipke, P.N. Role of force-sensitive amyloid-like interactions in fungal catch bonding and biofilms. Eukaryot. Cell 2014, 13, 1136–1142.
- Alsteens, D.; Garcia, M.C.; Lipke, P.N.; Dufrene, Y.F. Force-induced formation and propagation of adhesion nanodomains in living fungal cells. Proc. Natl. Acad. Sci. USA 2010, 107, 20744–20749.
- Formosa, C.; Schiavone, M.; Boisrame, A.; Richard, M.L.; Duval, R.E.; Dague, E. Multiparametric imaging of adhesive nanodomains at the surface of Candida albicans by atomic force microscopy. Nanomedicine 2015, 11, 57–65.
- de Groot, P.W.; Bader, O.; de Boer, A.D.; Weig, M.; Chauhan, N. Adhesins in human fungal pathogens: Glue with plenty of stick. Eukaryot. Cell 2013, 12, 470–481.
- Ho, V.; Herman-Bausier, P.; Shaw, C.; Conrad, K.A.; Garcia-Sherman, M.C.; Draghi, J.; Dufrene, Y.F.; Lipke, P.N.; Rauceo, J.M. An amyloid core sequence in the major candida albicans adhesin als1p mediates cell-cell adhesion. mBio 2019, 10, e01766-19.
- Lipke, P.N.; Mathelie-Guinlet, M.; Viljoen, A.; Dufrene, Y.F. A new function for amyloid-like interactions: Cross-beta aggregates of adhesins form cell-to-cell bonds. Pathogens 2021, 10, 1013.
- Schiavone, M.; Sieczkowski, N.; Castex, M.; Dague, E.; Marie Francois, J. Effects of the strain background and autolysis process on the composition and biophysical properties of the cell wall from two different industrial yeasts. FEMS Yeast Res. 2015, 15.
- Dehullu, J.; Valotteau, C.; Herman-Bausier, P.; Garcia-Sherman, M.; Mittelviefhaus, M.; Vorholt, J.A.; Lipke, P.N.; Dufrene, Y.F. Fluidic force microscopy demonstrates that homophilic adhesion by candida albicans ALS proteins is mediated by amyloid bonds between cells. Nano Lett. 2019, 19, 3846–3853.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
686
Revisions:
2 times
(View History)
Update Date:
29 Mar 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




