
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Cheng Zhang | + 2202 word(s) | 2202 | 2021-11-10 03:46:35 | | | |
| 2 | Lindsay Dong | + 310 word(s) | 2512 | 2021-12-13 03:36:36 | | |
Video Upload Options
Deoxyribonucleic acid (DNA), a genetic material, encodes all living information and living characteristics, e.g., in cell, DNA signaling circuits control the transcription activities of specific genes. In recent years, various DNA circuits have been developed to implement a wide range of signaling and for regulating gene network functions. In particular, a synthetic DNA circuit, with a programmable design and easy construction, has become a crucial method through which to simulate and regulate DNA signaling networks. Importantly, the construction of a hierarchical DNA circuit provides a useful tool for regulating gene networks and for processing molecular information. Moreover, via their robust and modular properties, DNA circuits can amplify weak signals and establish programmable cascade systems, which are particularly suitable for the applications of biosensing and detecting. Furthermore, a biological enzyme can also be used to provide diverse circuit regulation elements.
1. Introduction
Recently, various artificial DNA circuits have been established and widely applied to many fields such as medical diagnosis [1][2][3], molecular detection, and information processing [4][5][6][7][8][9][10]. Particularly, synthetic DNA circuits, designed and constructed in vitro, perform an important role in effectively controlling the gene networks in cell [11][12][13]. Synthetic DNA circuits have been demonstrated as possessing superiority in simulating and regulating DNA signaling, due to the properties of programmability and easy operation [14][15][16][17][18]. More importantly, synthetic DNA circuits have the potential to promote complex biological information processes and provide a new way to achieve gene analysis and molecular information processing [19][20][21].
Using predesigned specific base pair recognition, synthetic DNA circuits can modulate complex gene networks to implement diverse biofunctions. Recently, a variety of bioengineering and biocomputing functions have been regulated by varying the architectures and integrations of DNA circuits, such as their signal simulation [5][22], the molecular switch, catalytic cycle, cascade amplification [23][24][25][26][27][28][29][30], and logic gates [31][32][33][34][35][36][37]. In fact, most of the DNA circuits are implemented and regulated for the DNA strand displacement, whereby the longer DNA strand is able to hybridize with the complementary strand to displace the shorter one [38][39][40]. Through a DNA strand displacement reaction (SDR), a synthetic DNA circuit can be used to precisely regulate complex gene networks and molecular biosystems, e.g., DNA neural network systems that are constructed to implement pattern recognition [41].
2. The Research Studies on DNA Circuits
2.1. Cascading DNA Circuits In Vitro
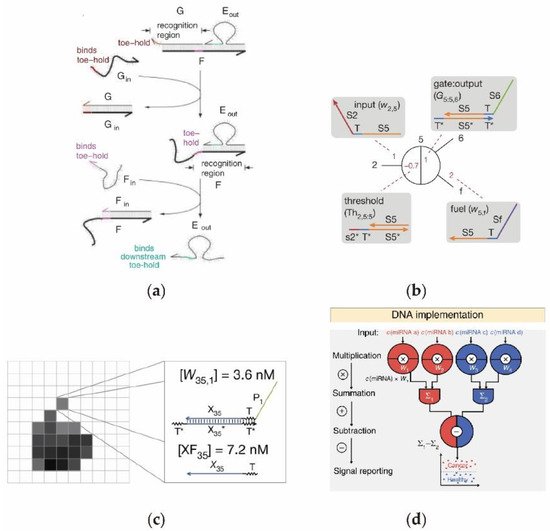
2.2. Enzyme-Free Catalytic DNA Circuit
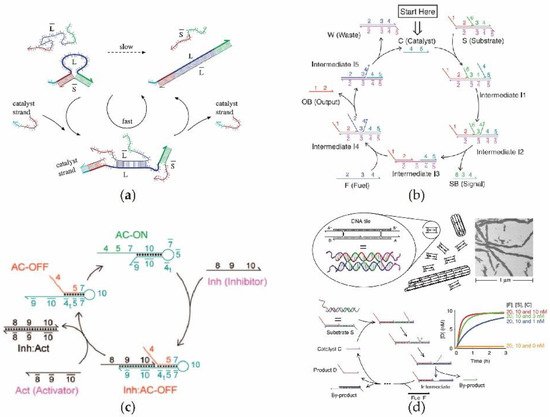
2.3. DNAzyme-Based DNA Circuit
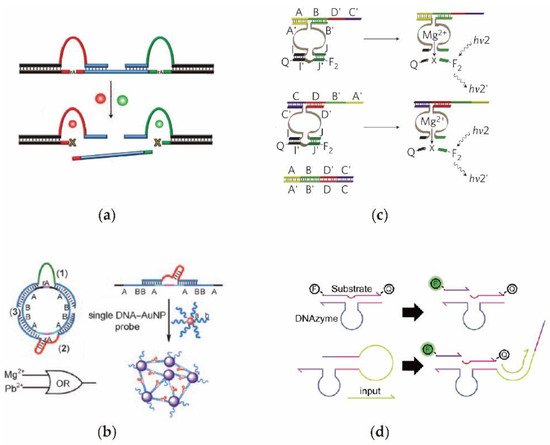
2.4. Protein Enzyme-Assiste d DNA Circuit
2.5. The DNA Circuits on Origami Surface
2.6. The DNA Circuits Combined with Nanoparticles
3. Summary
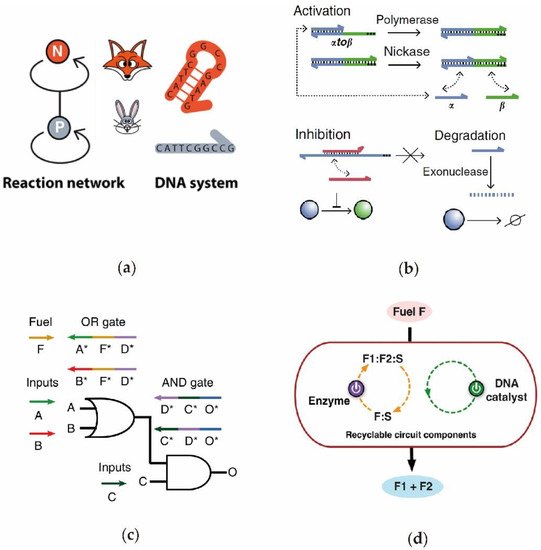
Although DNA circuits have engendered remarkable progresses in recent years, there are still some critical aspects that need to be improved upon to broaden the related application fields. The most important aspect is the signal leakage and distortion that exists in various complex DNA circuits [41][46]. The primary reason for this phenomenon is that some redundant base complementary pairings easily emerged during the circuit operations. At present, the design methods implemented to program DNA sequences are mainly based on the combinations of a manual design and simple software. Therefore, it is still difficult to completely eliminate these interference signals using pre-established methods, and a more automatic and universal sequence design platform is eagerly desired in the future. In addition, another important consideration to account for is the method by which to ensure the DNA circuit is reusable, thereby promoting its practical applications. Compared with the repeated electronic circuits, DNA circuits cannot be reused after completing the first complete operation [70][47][97], which inhibits the subsequent development of more advanced intelligent DNA circuits. Specifically, in large computing DNA circuits, it is important to establish a reusable circuit mechanism, to meet requirements of frequently adjusted circuit parameters and to test the structural stability of the system. In fact, many studies have been undertaken to improve the repeatability of the DNA circuits. More recently, Zhang et al. reported a nicking-assisted recycling strategy for implement reusable DNA circuits to achieve a repeatability of over 20 times during one reaction cycle [71].
References
- Du, Y.C.; Cui, Y.X.; Li, X.Y.; Sun, G.Y.; Zhang, Y.P.; Tang, A.N.; Kim, K.; Kong, D.M. Terminal Deoxynucleotidyl Transferase and T7 Exonuclease-Aided Amplification Strategy for Ultrasensitive Detection of Uracil-DNA Glycosylase. Anal. Chem. 2018, 90, 8629–8634.
- Shen, H.; Wang, J.; Liu, H.; Li, Z.; Jiang, F.; Wang, F.B.; Yuan, Q. Rapid and Selective Detection of Pathogenic Bacteria in Bloodstream Infections with Aptamer-Based Recognition. ACS Appl. Mater. Interfaces 2016, 8, 19371–19378.
- Zhang, Q.; Jiang, Q.; Li, N.; Dai, L.; Liu, Q.; Song, L.; Wang, J.; Li, Y.; Tian, J.; Ding, B.; et al. DNA Origami as an In Vivo Drug Delivery Vehicle for Cancer Therapy. ACS Nano 2014, 8, 6633–6643.
- Organick, L.; Ang, S.D.; Chen, Y.J.; Lopez, R.; Yekhanin, S.; Makarychev, K.; Racz, M.Z.; Kamath, G.; Gopalan, P.; Nguyen, B.; et al. Random access in large-scale DNA data storage. Nat. Biotechnol. 2018, 36, 242–248.
- Fujii, T.; Rondelez, Y. Predator-Prey Molecular Ecosystems. Acs Nano 2013, 7, 27–34.
- Guo, M.Y.; Chang, W.L.; Ho, M.C.; Lu, J.; Cao, J.N. Is optimal solution of every NP-complete or NP-hard problem determined from its characteristic for DNA-based computing. Biosystems 2005, 80, 71–82.
- Zhou, C.; Duan, X.Y.; Liu, N. A plasmonic nanorod that walks on DNA origami. Nat. Commun. 2015, 6, 6.
- Kim, J.; White, K.S.; Winfree, E. Construction of an in vitro bistable circuit from synthetic transcriptional switches. Mol. Syst. Biol. 2006, 2, 68.
- Xie, Z.; Liu, S.J.; Bleris, L.; Benenson, Y. Logic integration of mRNA signals by an RNAi-based molecular computer. Nucleic Acids Res. 2010, 38, 2692–2701.
- Wang, K.; Ren, J.; Fan, D.; Liu, Y.; Wang, E. Integration of graphene oxide and DNA as a universal platform for multiple arithmetic logic units. Chem. Commun. 2014, 50, 14390–14393.
- Zhu, J.B.; Zhang, L.B.; Dong, S.J.; Wang, E.K. Four-Way Junction-Driven DNA Strand Displacement and Its Application in Building Majority Logic Circuit. Acs Nano 2013, 7, 10211–10217.
- Fern, J.; Schulman, R. Design and Characterization of DNA Strand-Displacement Circuits in Serum-Supplemented Cell Medium. ACS Synth. Biol. 2017, 6, 1774–1783.
- Wei, Q.; Huang, J.; Li, J.; Wang, J.; Yang, X.; Liu, J.; Wang, K. A DNA nanowire based localized catalytic hairpin assembly reaction for microRNA imaging in live cells. Chem. Sci. 2018, 9, 7802–7808.
- Prokup, A.; Hemphill, J.; Deiters, A. DNA Computation: A Photochemically Controlled AND Gate. J. Am. Chem. Soc. 2012, 134, 3810–3815.
- Green, A.A.; Kim, J.; Ma, D.; Silver, P.A.; Collins, J.J.; Yin, P. Complex cellular logic computation using ribocomputing devices. Nature 2017, 548, 117–121.
- Rinaudo, K.; Bleris, L.; Maddamsetti, R.; Subramanian, S.; Weiss, R.; Benenson, Y. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat. Biotechnol. 2007, 25, 795–801.
- Hemphill, J.; Deiters, A. DNA computation in mammalian cells: microRNA logic operations. J. Am. Chem. Soc. 2013, 135, 10512–10518.
- Zong, Y.; Zhang, H.M.; Lyu, C.; Ji, X.; Hou, J.; Guo, X.; Ouyang, Q.; Lou, C. Insulated transcriptional elements enable precise design of genetic circuits. Nat. Commun. 2017, 8, 52.
- Phillips, A.; Cardelli, L. A programming language for composable DNA circuits. J. R. Soc. Interface 2009, 6 (Suppl. S4), 18.
- Chen, K.; Kong, J.; Zhu, J.; Ermann, N.; Predki, P.; Keyser, U.F. Digital Data Storage Using DNA Nanostructures and Solid-State Nanopores. Nano Lett. 2019, 19, 1210–1215.
- Lopez, R.; Chen, Y.J.; Dumas Ang, S.; Yekhanin, S.; Makarychev, K.; Racz, M.Z.; Seelig, G.; Strauss, K.; Ceze, L. DNA assembly for nanopore data storage readout. Nat. Commun. 2019, 10, 2933.
- Fern, J.; Scalise, D.; Cangialosi, A.; Howie, D.; Potters, L.; Schulman, R. DNA Strand-Displacement Timer Circuits. ACS Synth. Biol. 2017, 6, 190–193.
- Zhang, D.Y.; Winfree, E. Dynamic allosteric control of noncovalent DNA catalysis reactions. J. Am. Chem. Soc. 2008, 130, 13921–13926.
- Graugnard, E.; Kellis, D.L.; Bui, H.; Barnes, S.; Kuang, W.; Lee, J.; Hughes, W.L.; Knowlton, W.B.; Yurke, B. DNA-Controlled Excitonic Switches. Nano Lett. 2012, 12, 2117–2122.
- Chen, X.; Briggs, N.; McLain, J.R.; Ellington, A.D. Stacking nonenzymatic circuits for high signal gain. Proc. Natl. Acad. Sci. USA 2013, 110, 5386–5391.
- Wang, F.; Elbaz, J.; Willner, I. Enzyme-free amplified detection of DNA by an autonomous ligation DNAzyme machinery. J. Am. Chem. Soc. 2012, 134, 5504–5507.
- Wang, S.; Yue, L.; Shpilt, Z.; Cecconello, A.; Kahn, J.S.; Lehn, J.M.; Willner, I. Controlling the Catalytic Functions of DNAzymes within Constitutional Dynamic Networks of DNA Nanostructures. J. Am. Chem. Soc. 2017, 139, 9662–9671.
- Tian, Y.; Mao, C. Molecular gears: A pair of DNA circles continuously rolls against each other. J. Am. Chem. Soc. 2004, 126, 11410–11411.
- Wang, F.; Elbaz, J.; Orbach, R.; Magen, N.; Willner, I. Amplified analysis of DNA by the autonomous assembly of polymers consisting of DNAzyme wires. J. Am. Chem. Soc. 2011, 133, 17149–17151.
- Wang, F.; Elbaz, J.; Teller, C.; Willner, I. Amplified detection of DNA through an autocatalytic and catabolic DNAzyme-mediated process. Angew. Chem. 2011, 50, 295–299.
- Zhong, W.; Tang, W.; Tan, Y.; Fan, J.; Huang, Q.; Zhou, D.; Hong, W.; Liu, Y. A DNA arithmetic logic unit for implementing data backtracking operations. Chem. Commun. 2019, 55, 842–845.
- Shlyahovsky, B.; Li, Y.; Lioubashevski, O.; Elbaz, J.; Willner, I. Logic Gates and Antisense DNA Devices Operating on a Translator Nucleic Acid Scaffold. ACS Nano 2009, 3, 1831–1843.
- Lake, A.; Shang, S.; Kolpashchikov, D.M. Molecular Logic Gates Connected through DNA Four-Way Junctions. Angew. Chem. 2010, 49, 4459–4462.
- Li, W.; Zhang, F.; Yan, H.; Liu, Y. DNA based arithmetic function: A half adder based on DNA strand displacement. Nanoscale 2016, 8, 3775–3784.
- Lin, H.Y.; Chen, J.Z.; Li, H.Y.; Yang, C.N. A simple three-input DNA-based system works as a full-subtractor. Sci. Rep. 2015, 5, 10686.
- Wu, C.; Wang, K.; Fan, D.; Zhou, C.; Liu, Y.; Wang, E. Enzyme-free and DNA-based multiplexer and demultiplexer. Chem. Commun. 2015, 51, 15940–15943.
- Zhu, J.; Zhang, L.; Li, T.; Dong, S.; Wang, E. Enzyme-Free Unlabeled DNA Logic Circuits Based on Toehold-Mediated Strand Displacement and Split G-Quadruplex Enhanced Fluorescence. Adv. Mater. 2013, 25, 2440–2444.
- Chen, X. Expanding the rule set of DNA circuitry with associative toehold activation. J. Am. Chem. Soc. 2012, 134, 263–271.
- Zhang, D.Y.; Winfree, E. Control of DNA Strand Displacement Kinetics Using Toehold Exchange. J. Am. Chem. Soc. 2009, 131, 17303–17314.
- Xing, Y.; Yang, Z.; Liu, D. A Responsive Hidden Toehold To Enable Controllable DNA Strand Displacement Reactions. Angew. Chem. 2011, 50, 11934–11936.
- Cherry, K.M.; Qian, L. Scaling up molecular pattern recognition with DNA-based winner-take-all neural networks. Nature 2018, 559, 370–376.
- Chatterjee, G.; Dalchau, N.; Muscat, R.A.; Phillips, A.; Seelig, G. A spatially localized architecture for fast and modular DNA computing. Nat. Nanotechnol. 2017, 12, 920–927.
- Srinivas, N.; Ouldridge, T.E.; Sulc, P.; Schaeffer, J.M.; Yurke, B.; Louis, A.A.; Doye, J.P.; Winfree, E. On the biophysics and kinetics of toehold-mediated DNA strand displacement. Nucleic Acids Res. 2013, 41, 10641–10658.
- Frezza, B.M.; Cockroft, S.L.; Ghadiri, M.R. Modular multi-level circuits from immobilized DNA-Based logic gates. J. Am. Chem. Soc. 2007, 129, 14875–14879.
- Seelig, G.; Soloveichik, D.; Zhang, D.Y.; Winfree, E. Enzyme-free nucleic acid logic circuits. Science 2006, 314, 1585–1588.
- Qian, L.; Winfree, E. Scaling Up Digital Circuit Computation with DNA Strand Displacement Cascades. Science 2011, 332, 1196–1201.
- Zhang, C.; Zhao, Y.; Xu, X.; Xu, R.; Li, H.; Teng, X.; Du, Y.; Miao, Y.; Lin, H.C.; Han, D. Cancer diagnosis with DNA molecular computation. Nat. Nanotechnol. 2020, 15, 709–715.
- Green, S.J.; Lubrich, D.; Turberfield, A.J. DNA hairpins: Fuel for autonomous DNA devices. Biophys. J. 2006, 91, 2966–2975.
- Seelig, G.; Yurke, B.; Winfree, E. Catalyzed relaxation of a metastable DNA fuel. J. Am. Chem. Soc. 2006, 128, 12211–12220.
- Zhang, D.Y.; Turberfield, A.J.; Yurke, B.; Winfree, E. Engineering entropy-driven reactions and networks catalyzed by DNA. Science 2007, 318, 1121–1125.
- Zhang, D.Y.; Hariadi, R.F.; Choi, H.M.; Winfree, E. Integrating DNA strand-displacement circuitry with DNA tile self-assembly. Nat. Commun. 2013, 4, 1965.
- Elbaz, J.; Moshe, M.; Shlyahovsky, B.; Willner, I. Cooperative multicomponent self-assembly of nucleic acid structures for the activation of DNAzyme cascades: A paradigm for DNA sensors and aptasensors. Chemistry 2009, 15, 3411–3418.
- Orbach, R.; Mostinski, L.; Wang, F.; Willner, I. Nucleic acid driven DNA machineries synthesizing Mg2+-dependent DNAzymes: An interplay between DNA sensing and logic-gate operations. Chemistry 2012, 18, 14689–14694.
- Huang, P.J.; Lin, J.; Cao, J.; Vazin, M.; Liu, J. Ultrasensitive DNAzyme beacon for lanthanides and metal speciation. Anal. Chem. 2014, 86, 1816–1821.
- Wu, N.; Willner, I. DNAzyme-Controlled Cleavage of Dimer and Trimer Origami Tiles. Nano Lett. 2016, 16, 2867–2872.
- Endo, M.; Takeuchi, Y.; Suzuki, Y.; Emura, T.; Hidaka, K.; Wang, F.; Willner, I.; Sugiyama, H. Single-Molecule Visualization of the Activity of a Zn(2+)-Dependent DNAzyme. Angew. Chem. 2015, 54, 10550–10554.
- Aleman-Garcia, M.A.; Orbach, R.; Willner, I. Ion-responsive hemin-G-quadruplexes for switchable DNAzyme and enzyme functions. Chem. 2014, 20, 5619–5624.
- Wang, F.; Orbach, R.; Willner, I. Detection of metal ions (Cu2+, Hg2+) and cocaine by using ligation DNAzyme machinery. Chem. 2012, 18, 16030–16036.
- Yang, J.; Wu, R.; Li, Y.; Wang, Z.; Pan, L.; Zhang, Q.; Lu, Z.; Zhang, C. Entropy-driven DNA logic circuits regulated by DNAzyme. Nucleic Acids Res. 2018, 46, 8532–8541.
- Orbach, R.; Willner, B.; Willner, I. Catalytic nucleic acids (DNAzymes) as functional units for logic gates and computing circuits: From basic principles to practical applications. Chem. Commun. 2015, 51, 4144–4160.
- Moshe, M.; Elbaz, J.; Willner, I. Sensing of UO22+ and Design of Logic Gates by the Application of Supramolecular Constructs of Ion-Dependent DNAzymes. Nano Lett. 2009, 9, 1196–1200.
- Bi, S.; Yan, Y.; Hao, S.; Zhang, S. Colorimetric logic gates based on supramolecular DNAzyme structures. Angew. Chem. 2010, 49, 4438–4442.
- Elbaz, J.; Lioubashevski, O.; Wang, F.; Remacle, F.; Levine, R.D.; Willner, I. DNA computing circuits using libraries of DNAzyme subunits. Nat. Nanotechnol. 2010, 5, 417–422.
- Harding, B.I.; Pollak, N.M.; Stefanovic, D.; Macdonald, J. Repeated Reuse of Deoxyribozyme-Based Logic Gates. Nano Lett. 2019, 19, 7655–7661.
- Enghiad, B.; Zhao, H.M. Programmable DNA-Guided Artificial Restriction Enzymes. ACS Synth. Biol. 2017, 6, 752–757.
- Yang, X.L.; Tang, Y.A.; Mason, S.D.; Chen, J.B.; Li, F. Enzyme-Powered Three-Dimensional DNA Nanomachine for DNA Walking, Payload Release, and Biosensing. Acs Nano 2016, 10, 2324–2330.
- Wang, F.; Zahid, O.K.; Swain, B.E.; Parsonage, D.; Hollis, T.; Harvey, S.; Perrino, F.W.; Kohli, R.M.; Taylor, E.W.; Hall, A.R. Solid-State Nanopore Analysis of Diverse DNA Base Modifications Using a Modular Enzymatic Labeling Process. Nano Lett. 2017, 17, 7110–7116.
- Esadze, A.; Rodriguez, G.; Weiser, B.P.; Cole, P.A.; Stivers, J.T. Measurement of nanoscale DNA translocation by uracil DNA glycosylase in human cells. Nucleic Acids Res. 2017, 45, 12413–12424.
- Meijer, L.; Joesaar, A.; Steur, E.; Engelen, W.; van Santen, R.A.; Merkx, M.; de Greef, T. Hierarchical control of enzymatic actuators using DNA-based switchable memories. Nat. Commun. 2017, 8, 11.
- Song, T.; Eshra, A.; Shah, S.; Bui, H.; Fu, D.; Yang, M.; Mokhtar, R.; Reif, J. Fast and compact DNA logic circuits based on single-stranded gates using strand-displacing polymerase. Nat. Nanotechnol. 2019, 14, 1075–1081.
- Zhang, C.; Wang, Z.; Liu, Y.; Yang, J.; Zhang, X.; Li, Y.; Pan, L.; Ke, Y.; Yan, H. Nicking-Assisted Reactant Recycle To Implement Entropy-Driven DNA Circuit. J. Am. Chem. Soc. 2019, 141, 17189–17197.
- Johnson, J.A.; Dehankar, A.; Winter, J.O.; Castro, C.E. Reciprocal Control of Hierarchical DNA Origami-Nanoparticle Assemblies. Nano Lett. 2019, 19, 8469–8475.
- Wang, J.; Yue, L.; Li, Z.; Zhang, J.; Tian, H.; Willner, I. Active generation of nanoholes in DNA origami scaffolds for programmed catalysis in nanocavities. Nat. Commun. 2019, 10, 4963.
- Tikhomirov, G.; Petersen, P.; Qian, L. Triangular DNA Origami Tilings. J. Am. Chem. Soc. 2018, 140, 17361–17364.
- Tian, Y.; Wang, T.; Liu, W.; Xin, H.L.; Li, H.; Ke, Y.; Shih, W.M.; Gang, O. Prescribed nanoparticle cluster architectures and low-dimensional arrays built using octahedral DNA origami frames. Nat. Nanotechnol. 2015, 10, 637–644.
- Castro, C.E.; Kilchherr, F.; Kim, D.N.; Shiao, E.L.; Wauer, T.; Wortmann, P.; Bathe, M.; Dietz, H. A primer to scaffolded DNA origami. Nat. Methods 2011, 8, 221–229.
- Chao, J.; Wang, J.; Wang, F.; Ouyang, X.; Kopperger, E.; Liu, H.; Li, Q.; Shi, J.; Wang, L.; Hu, J.; et al. Solving mazes with single-molecule DNA navigators. Nat. Mater. 2019, 18, 273–279.
- Wickham, S.F.; Bath, J.; Katsuda, Y.; Endo, M.; Hidaka, K.; Sugiyama, H.; Turberfield, A.J. A DNA-based molecular motor that can navigate a network of tracks. Nat. Nanotechnol. 2012, 7, 169–173.
- Teichmann, M.; Kopperger, E.; Simmel, F.C. Robustness of Localized DNA Strand Displacement Cascades. ACS Nano 2014, 8, 8487–8496.
- Thubagere, A.J.; Li, W.; Johnson, R.F.; Chen, Z.; Doroudi, S.; Lee, Y.L.; Izatt, G.; Wittman, S.; Srinivas, N.; Woods, D.; et al. A cargo-sorting DNA robot. Science 2017, 357, 9.
- Chen, G.; Gibson, K.J.; Liu, D.; Rees, H.C.; Lee, J.H.; Xia, W.; Lin, R.; Xin, H.L.; Gang, O.; Weizmann, Y. Regioselective surface encoding of nanoparticles for programmable self-assembly. Nat. Mater. 2019, 18, 169–174.
- Lan, X.; Lu, X.; Shen, C.; Ke, Y.; Ni, W.; Wang, Q. Au nanorod helical superstructures with designed chirality. J. Am. Chem. Soc. 2015, 137, 457–462.
- Lan, X.; Liu, T.; Wang, Z.; Govorov, A.O.; Yan, H.; Liu, Y. DNA-Guided Plasmonic Helix with Switchable Chirality. J. Am. Chem. Soc. 2018, 140, 11763–11770.
- Thacker, V.V.; Herrmann, L.O.; Sigle, D.O.; Zhang, T.; Liedl, T.; Baumberg, J.J.; Keyser, U.F. DNA origami based assembly of gold nanoparticle dimers for surface-enhanced Raman scattering. Nat. Commun. 2014, 5.
- Kühler, P.; Roller, E.M.; Schreiber, R.; Liedl, T.; Lohmüller, T.; Feldmann, J. Plasmonic DNA-Origami Nanoantennas for Surface-Enhanced Raman Spectroscopy. Nano Lett. 2014, 14, 2914–2919.
- Xin, L.; Lu, M.; Both, S.; Pfeiffer, M.; Urban, M.J.; Zhou, C. Watching a Single Fluorophore Molecule Walk into a Plasmonic Hotspot. Acs Photonics 2019, 6, 985–993.
- Urban, M.J.; Zhou, C.; Duan, X.; Liu, N. Optically Resolving the Dynamic Walking of a Plasmonic Walker Couple. Nano Lett. 2015, 15, 8392–8396.
- Kyriazi, M.E.; Giust, D.; El-Sagheer, A.H.; Lackie, P.M.; Muskens, O.L.; Brown, T.; Kanaras, A.G. Multiplexed mRNA Sensing and Combinatorial-Targeted Drug Delivery Using DNA-Gold Nanoparticle Dimers. Acs Nano 2018, 12, 3333–3340.
- Chai, H.; Miao, P. Bipedal DNA Walker Based Electrochemical Genosensing Strategy. Anal. Chem. 2019, 91, 4953–4957.
- Chen, Y.; Corn, R.M. DNAzyme footprinting: Detecting protein-aptamer complexation on surfaces by blocking DNAzyme cleavage activity. J. Am. Chem. Soc. 2013, 135, 2072–2075.
- Jiang, X.; Wang, H.; Chai, Y.; Li, H.; Shi, W.; Yuan, R. DNA Cascade Reaction with High-Efficiency Target Conversion for Ultrasensitive Electrochemiluminescence microRNA Detection. Anal. Chem. 2019, 91, 10258–10265.
- Zhou, J.; Lai, W.; Zhuang, J.; Tang, J.; Tang, D. Nanogold-functionalized DNAzyme concatamers with redox-active intercalators for quadruple signal amplification of electrochemical immunoassay. ACS Appl. Mater. Interfaces 2013, 5, 2773–2781.
- Guo, X.; Li, F.; Bai, L.; Yu, W.; Zhang, X.; Zhu, Y.; Yang, D. Gene Circuit Compartment on Nanointerface Facilitatating Cascade Gene Expression. J. Am. Chem. Soc. 2019, 141, 19171–19177.
- Yang, Y.; Huang, J.; Yang, X.; He, X.; Quan, K.; Xie, N.; Ou, M.; Wang, K. Gold Nanoparticle Based Hairpin-Locked-DNAzyme Probe for Amplified miRNA Imaging in Living Cells. Anal. Chem. 2017, 89, 5850–5856.
- Liang, C.P.; Ma, P.Q.; Liu, H.; Guo, X.; Yin, B.C.; Ye, B.C. Rational Engineering of a Dynamic, Entropy-Driven DNA Nanomachine for Intracellular MicroRNA Imaging. Angew. Chem. 2017, 56, 9077–9081.
- Gines, G.; Zadorin, A.S.; Galas, J.C.; Fujii, T.; Estevez-Torres, A.; Rondelez, Y. Microscopic agents programmed by DNA circuits. Nat. Nanotechnol. 2017, 12, 351–359.
- Yao, G.; Li, J.; Li, Q.; Chen, X.; Liu, X.; Wang, F.; Qu, Z.; Ge, Z.; Narayanan, R.P.; Williams, D.; et al. Programming nanoparticle valence bonds with single-stranded DNA encoders. Nat. Mater. 2019, 19, 781–788.





