Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Sadanand Pandey | + 2227 word(s) | 2227 | 2021-11-18 06:47:54 | | | |
| 2 | Lindsay Dong | + 7 word(s) | 2234 | 2021-12-13 04:01:46 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Pandey, S. Nanotechnology-Based Techniques for Sensing Amino Acids/Peptides/Proteins. Encyclopedia. Available online: https://encyclopedia.pub/entry/16886 (accessed on 07 March 2026).
Pandey S. Nanotechnology-Based Techniques for Sensing Amino Acids/Peptides/Proteins. Encyclopedia. Available at: https://encyclopedia.pub/entry/16886. Accessed March 07, 2026.
Pandey, Sadanand. "Nanotechnology-Based Techniques for Sensing Amino Acids/Peptides/Proteins" Encyclopedia, https://encyclopedia.pub/entry/16886 (accessed March 07, 2026).
Pandey, S. (2021, December 08). Nanotechnology-Based Techniques for Sensing Amino Acids/Peptides/Proteins. In Encyclopedia. https://encyclopedia.pub/entry/16886
Pandey, Sadanand. "Nanotechnology-Based Techniques for Sensing Amino Acids/Peptides/Proteins." Encyclopedia. Web. 08 December, 2021.
Copy Citation
Protein, peptide- and amino acid-based drug delivery systems have proficiently transformed nanotechnology via immense flexibility in their features for attaching various drug molecules and biodegradable polymers. In this regard, novel nanostructures including carbon nanotubes, electrospun carbon nanofibers, gold nanoislands, and metal-based nanoparticles have been introduced as nanosensors for accurate detection of these organic compounds. These nanostructures can bind the biological receptor to the sensor surface and increase the surface area of the working electrode, significantly enhancing the biosensor performance.
amino acids
proteins
peptides
nanomaterials
detection
1. Routine Methods for Detection of AAs, Proteins, and Peptides
Proteins are complex molecules essential to life that have enzymatic, structural, and storage functions. The most common techniques used to determine the total amount of protein are isotope ratio mass spectrometry (IRMS), the Kjeldahl method [1], and biuret methods such as the Lowry′s method [2] and the Bradford method [3]. Among them, the IRMS and Kjeldahl methods are susceptible and reproducible. However, artifacts have been observed in these methods. The interference effect is relatively high in spectrophotometric and colorimetric techniques used to determine the total protein amount. Therefore, the desired protein must be purified in the first step. However, this results in the loss of some proteins. None of the abovementioned methods provides information about AA composition.
The importance of AA analysis is increasing daily in different fields such as biochemistry, clinical chemistry, nutrition, and pharmaceutical formulation. The AA contents, chemical forms, and sample matrices (food, biological fluid, or protein hydrolysis) of many samples are quite different. AAs play a significant role in forming vital biomolecules such as hormones, neurotransmitters, antibodies, and signaling molecules. Since AAs are the precursors of many biomarkers, determining the amount of AAs in biological fluids is essential for the early diagnosis of many diseases. Studies have reported that many AAs play a role in forming diseases such as phenylketonuria, citrullinemia, and homocystinuria diseases [4][5].
Determining the separation and amount of AAs is very important to provide information about polypeptides’ and proteins′ characterization and structural properties. However, these compounds are difficult to identify and separate because of their high polarity and lack of strong chromophoric groups. Since many commonly used AAs cannot be determined directly by spectroscopic methods (UV–visible spectrophotometry or fluorometry), the amino groups of AAs are selectively modified with substances that show fluorescence or visible-light absorption prior to their determination [6]. Mass spectrometry (MS) and chromatography combination are currently used as analysis platforms. The separation and quantitative analysis of free AAs before or after protein hydrolysis is carried out with the aid of modern methods such as ion-exchange chromatography (IEC), gas chromatography/mass spectrometry (GC/MS), and liquid chromatography-mass spectrometry/mass spectrometry (LC–MS/MS). Each method comes with its own advantages and disadvantages.
Using the GC/MS method instead of GC with flame ionization or electron capture makes AA analysis more attractive. GC provides short analysis times, but AAs need to be derivatized into GC-detectable forms. However, this process also prolongs the analysis time. Substances such as N,O-bis-(trimethylsilyl), trifluoroacetamide (BSTFA), or N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA) can be used for derivatization. Still, steric hindrance due to the formation of bulky groups can be developed [7]. In 1998, Husek described rapid derivatization (about 1 min) of AAs with alkyl chloroformates. In this method, the esterification of carboxylic acids, amino groups, and hydroxyl groups was carried out to form alkyl esters or N(O)-alkoxycarbonyl ethers, and AA analysis could be performed in less than 10 min [8][9].
Moore and Stein were the first to develop an IEC-based AA analyzer in the 1960s [10]. In today’s methods, IEC and gas/liquid chromatography techniques are applied using different detectors. IEC coupled to the postcolumn ninhydrin derivatization method is the most widely used technique in the clinical field. It is considered a gold standard for detecting AAs in biological samples because of its wide dynamic range and linearity. The major disadvantage is that it is a time-consuming method (usually 2–3 h per sample) that requires high sample volumes (>200 µL). In addition, detecting interfering compounds that react with ninhydrin and cannot be determined by spectrophotometric detection generates problems [11][12]. The LC-MS/MS technique has become a compelling tool because of its better selectivity and shorter analysis times compared to IEC. In 2018, Casado and coworkers aimed to develop an ultraperformance liquid chromatography–tandem mass spectrometry (UPLC–MS/MS) procedure to identify 25 AAs and 17 related compounds in plasma, urine, cerebrospinal fluid (CSF), and dried bloodstains. The comparison of the results obtained from this procedure with those derived from IEC revealed a good correlation between the two techniques except for 4-hydroxyproline, aspartate, and citrulline [12]. In 2020, Carling and coworkers investigated and compared the analytical performance of three commercially available reagent kits for LC–MS, IEC, and LC–MS/MS, used for plasma AA analysis. According to their results, the LC–MS test showed a low correlation with IEC, while LC–MS/MS showed a good correlation with IEC. It was stated that IEC should no longer be defined as the gold standard method for plasma AA analysis, as LC-MS/MS offered superior specificity and faster analysis time. Although the sensitivity of the chromatographic techniques is high, they are expensive, do not allow point-of-care analysis, and require killed personnel. Detection of proteins by direct protein electrochemistry makes them suitable for ‘point of care’ or ‘in-field testing’ applications. Also, the electrochemistry of direct protein enables the detection of conformational changes and modifications in proteins [13].
2. Different Nanomaterials as Nanosensors for Detecting AAs, Proteins, and Peptides
Nanomaterials are promising materials with at least one size in the range of 1–100 nm. Outstandingly high surface areas can be attained via the intelligent design of nanomaterials. Furthermore, nanomaterials can be synthesized with outstanding electrical, optical, catalytic, and mechanical properties that are superior to those of their bulk counterparts. Nanomaterial properties can be adjusted as desired via controlling the synthesis conditions and adequate functionalization [14].
Nanomaterials can be categorized into three classes according to their content: (i) organic-carbon-based nanomaterials-carbon nanotubes (CNTs), carbon nanofibers (CNFs), fullerenes (C60), and graphene (GR). Chemical vapor deposition (CVD) [15], laser ablation [16][17], and arc discharge techniques [18][19] are used for the production of organic-carbon-based nanomaterials; (ii) inorganic-based nanomaterials—quantum dots, gold NPs, and magnetic NPs. These nanomaterials can be synthesized into metals such as Au or AgNPs, metal oxides such as TiO2 and ZnO NPs, and semiconductors such as silicon and ceramics; (iii) hybrid nanomaterials, which can be any combination of carbon-based, metal-based, or organic-based nanomaterials with any form of metal, ceramic, or polymer bulk materials [20].
A sensor is an analytical device that can detect and quantify the presence of an analyte in a sample. It includes receptors, transducers, and reading systems. The biological receptor interacts specifically with the target analyte, and the transducer converts this information into a measurable signal [21]. For example, piezoelectric transducers are involved in measuring the change in mass after the formation of analyte–bioreceptor complexes, while optical transducers and electrochemical transducers measure the changes in light intensity and conductivity, current, or potential, respectively. Finally, the magnitude of the change is measured by the reading system. Figure 1 shows a schematic diagram of a typical biosensor.
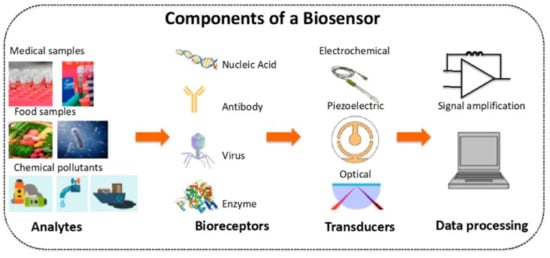
Figure 1. Schematic diagram of a typical biosensor. Reprinted with permission from ref. [22].
Bio-based analysis systems have recently become the most used and desired devices for diagnosis in the clinical field because of their fast response times and reliable features. In biosensors, a biological element (the receptor) is immobilized on the transducer using different strategies [23]. Analyte detection is performed using the high affinity between the receptor and its ligands, such as antigen-antibody, enzymatic (enzyme–substrate), or cellular (microorganisms, proteins) interactions. The ability to detect important biomarkers such as nucleic acids, AAs, and proteins associated with a disease is essential for the clinical field [24]. The technique of immobilization of the biofunctional component on the working electrode dramatically affects the performance of biosensors. It is important to note that a biosensor’s stability is not lost while forming a close relationship between the biological component and the sensor surface (transducer). Therefore, the selection of immobilization matrices that support the performance of the sensor system is very critical.
Nanosensors are sensing devices with at least one sensing size smaller than 100 nm [25]. The use of nanoscale materials as reinforcement increases the interface area of the resulting composites. For this reason, various reinforcement elements such as hydroxyapatite, gold NPs, GR, CNTs, and CNFs are used to increase the surface area and especially the conductivity in sensor applications [26]. Carbon-based nanomaterials are widely used as reinforcements because of their stable, mechanically robust, flexible, electrical, and thermally conductive properties. Thus, these nanomaterials are promising in the development of high-performance devices [27].
Macro- and microscale sensors such as electrochemical and optical sensors are currently being used in the clinical field. For example, electrochemical and optical sensors such as blood gas and pH are frequently used in intensive care. Likewise, disposable electrodes are used in the clinical field to record biopotentials such as electrocardiograms and electroencephalograms [28]. Nevertheless, the use of nanosensors in the early-stage diagnosis of diseases and preclinical studies is increasing. In particular, whole-cell behaviors, adhesion processes of cells to the extracellular matrix, and cell-cell interactions can be easily monitored in vitro thanks to label-free electrochemical nanosensors [29]. For example, in vitro studies can be performed in the presence of components (drug or toxic substance) that can affect cell adhesions to the biofunctional surface of a nanosensor developed on a cell-based platform under the electrochemical measurements. This sheds light on the studies carried out before the transition to in vivo applications, which is the next step of preclinical studies. This also reduces animal experiments by using these developed nanosensors. At the same time, nanosensors are attracting much attention as an alternative to the invasive methods currently used to diagnose diseases in the clinical field. Recently developed wearable nanosensors are promising for noninvasive monitoring of biomarkers. It is crucial that some compounds that serve as disease biomarkers can be determined from saliva, sweat, or tears. At the same time, electrochemical nanosensors with increased stability are being developed for real-time monitoring of small molecules in blood or drug-active substances in plasma in a continuous flow environment [30].
2.1. Metal NP-Based Sensors
With the development of nanoscience and nanotechnology, metal NPs are highly desirable in areas such as nanosensors, biomedicine, biological labeling, and microelectronics because of their unique properties such as sizeable surface-to-volume ratio and high electrical conductivity, biocompatibility, catalytic activity, etc. [31]. Signal-generating molecules are usually used to bind bioreceptors to the biosensor recognition surface for labeling. Enzymes such as horseradish peroxidase are labeled agents and require an additional dye or substrate in affinity-based sensors. Enzyme labels are not stable, since they are affected by environmental conditions. Additionally, they are expensive. Nanoprobes have become quite popular as an alternative. Usage of electroactive NPs as nanolabels contributes to improving biosensor performance. Furthermore, electroactive NPs are inexpensive and stable [32].
2.2. Carbon-Based Nanomaterials
Carbon-based nanomaterials display outstanding properties such as high electrical conductivity, fast electron transfer capability, and high specific surface area, making them highly interesting for developing high-performance biosensors [33]. Commonly used carbon nanomaterials are carbon nanotubes (CNTs) and graphene and its derivatives, in the forms of nanotubes and platelets, respectively.
2.3. Electrospun Nanofibers (ESNFs)
Electrospinning is defined as the production of nanofibers from polymer solutions under a high electric field (kV) [34]. It is the only method for mass production of continuous long nanofibers [35]. Among the numerous nanomaterials, ESNFs are building materials in drug delivery systems, biosensors, biomedicine, food textile, and environmental applications because of their large surface areas, controllable surface conformations, porous structures, and high concentrations adsorption capacity, and good biocompatibility [36][37][38]. Because of these properties, electrospun nanofibers have better sensitivity than sensors formed with other materials. In addition, biomimetic coatings can prevent biofouling, thereby extending the life of biosensors [39]. ESNFs are produced via electrospinning, which is a simple, effective, controlled, and economical method. Fibers can be obtained from various materials; solutions or melt forms of organic polymers are among the most common sources. In particular, the production of nanofibers is possible from composite materials obtained by the appropriate combination of components with different morphologies in the nano size (e.g., NPs, nanorods, nanowires, nanotubes, and nanosheets) with organic polymers. Figure 2 shows a schematic representation of a conventional electrospinning setup.
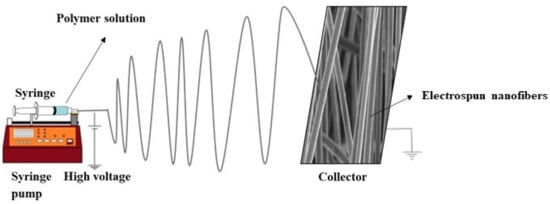
Figure 2. Representation of a conventional electrospinning setup.
2.4. Molecularly Imprinted Polymers
Molecular imprinting is a promising method for developing affinity-based nanomaterials with high specific recognition ability [40][41]. Molecularly imprinted polymers (MIPs) provide many properties such as selectivity, stability, reusability, and low cost compared with biological recognition materials such as enzymes and antibodies. They have some drawbacks, such as a high diffusion barrier and low space accessibility, given that most of the imprinted areas are formed inside the MIP. To overcome these issues, the surface printing technique, which involves the production of a MIP layer on the surface of nanomaterials, has been developed in recent years. This method provides the advantages of higher bonding capacity and faster bonding kinetics on the material surface [42]. The applications of MIPs combined with electrochemical studies have increased in the sensor field because of their ease of use and low cost [43]. However, some problems still need to be overcome before MIP-based sensors can enter the sensor market. The most significant change is in the distance of the imprinted cavities to the sensor surface and, accordingly, low signal reception [44]. Therefore, researchers have focused on improving the surface of nanosized support materials such as GR with ultrathin polymeric films. Through this method, higher selectivity is provided for thin MIP layers [45].
References
- Gibson, R.B. The Determination of Nitrogen by the Kjeldahl Method. J. Am. Chem. Soc. 1904, 26, 105–110.
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275.
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254.
- Sandlers, Y. Amino acids profiling for the diagnosis of metabolic disorders. In Biochemical Testing-Clinical Correlation and Diagnosis; IntechOpen: London, UK, 2019.
- Gałęzowska, G.; Ratajczyk, J.; Wolska, L. Determination of amino acids in human biological fluids by high-performance liquid chromatography: Critical review. Amino Acids 2021, 53, 993–1009.
- Shinichi Ozawa, H. Advances in amino acid analysis and Amino Acid Analyzer L-8900. Hitachi Sci. Instrum. News 2015, 6, 33–43.
- Moldoveanu, S.C.; David, V. Derivatization methods in GC and GC/MS. In Gas Chromatography-Derivatization, Sample Preparation, Application; IntechOpen: London, UK, 2018.
- Hušek, P. Chloroformates in gas chromatography as general purpose derivatizing agents. J. Chromatogr. B Biomed. Sci. Appl. 1998, 717, 57–91.
- Zhao, L.; Ni, Y.; Su, M.; Li, H.; Dong, F.; Chen, W.; Wei, R.; Zhang, L.; Guiraud, S.P.; Martin, F.-P. High throughput and quantitative measurement of microbial metabolome by gas chromatography/mass spectrometry using automated alkyl chloroformate derivatization. Anal. Chem. 2017, 89, 5565–5577.
- Moore, S.; Stein, W.H. Chromatographic determination of amino acids by the use of automatic recording equipment. Methods Enzymol. 1963, 6, 819–831.
- Smon, A.; Cuk, V.; Brecelj, J.; Murko, S.; Groselj, U.; Tansek, M.Z.; Battelino, T.; Lampret, B.R. Comparison of liquid chromatography with tandem mass spectrometry and ion-exchange chromatography by post-column ninhydrin derivatization for amino acid monitoring. Clin. Chim. Acta 2019, 495, 446–450.
- Casado, M.; Sierra, C.; Batllori, M.; Artuch, R.; Ormazabal, A. A targeted metabolomic procedure for amino acid analysis in different biological specimens by ultra-high-performance liquid chromatography–tandem mass spectrometry. Metabolomics 2018, 14, 1–12.
- Carling, R.S.; McDonald, B.A.; Austin, D.; Burden, D.; Correia, J.; Leung, J.; Mayers, B.; John, C. Challenging the status quo: A comparison of ion exchange chromatography with liquid chromatography–mass spectrometry and liquid chromatography–tandem mass spectrometry methods for the measurement of amino acids in human plasma. Ann. Clin. Biochem. 2020, 57, 277–290.
- Baig, N.; Kammakakam, I.; Falath, W. Nanomaterials: A review of synthesis methods, properties, recent progress, and challenges. Mater. Adv. 2021, 2, 1821–1871.
- Paradise, M.; Goswami, T. Carbon nanotubes–production and industrial applications. Mater. Des. 2007, 28, 1477–1489.
- Scott, C.D.; Arepalli, S.; Nikolaev, P.; Smalley, R.E. Growth mechanisms for single-wall carbon nanotubes in a laser-ablation process. Appl. Phys. A 2001, 72, 573–580.
- Rafique, M.M.A.; Iqbal, J. Production of carbon nanotubes by different routes-a review. J. Encapsulation Adsorpt. Sci. 2011, 1, 29.
- Thostenson, E.T.; Ren, Z.; Chou, T.-W. Advances in the science and technology of carbon nanotubes and their composites: A review. Compos. Sci. Technol. 2001, 61, 1899–1912.
- Hosseini, A.; Allahyari, M.; Besheli, S.D. Synthesis of carbon nanotubes, nano fibbers and nano union by electric arc discharge method using NaCl accuse as solution and Fe and Ni particles and catalysts. IJEST 2012, 1, 217–229.
- Al-Kayiem, H.H.; Lin, S.C.; Lukmon, A. Review on nanomaterials for thermal energy storage technologies. Nanosci. Nanotechnol. Asia 2013, 3, 60–71.
- Grieshaber, D.; MacKenzie, R.; Vörös, J.; Reimhult, E. Electrochemical biosensors-sensor principles and architectures. Sensors 2008, 8, 1400–1458.
- Zhou, Y.; Fang, Y.; Ramasamy, R.P. Non-covalent functionalization of carbon nanotubes for electrochemical biosensor development. Sensors 2019, 19, 392.
- Sage, A.T.; Besant, J.D.; Lam, B.; Sargent, E.H.; Kelley, S.O. Ultrasensitive electrochemical biomolecular detection using nanostructured microelectrodes. Acc. Chem. Res. 2014, 47, 2417–2425.
- Abdel-Karim, R.; Reda, Y.; Abdel-Fattah, A. Nanostructured materials-based nanosensors. J. Electrochem. Soc. 2020, 167, 037554.
- Munawar, A.; Ong, Y.; Schirhagl, R.; Tahir, M.A.; Khan, W.S.; Bajwa, S.Z. Nanosensors for diagnosis with optical, electric and mechanical transducers. RSC Adv. 2019, 9, 6793–6803.
- Naskar, D.; Bhattacharjee, P.; Ghosh, A.K.; Mandal, M.; Kundu, S.C. Carbon nanofiber reinforced nonmulberry silk protein fibroin nanobiocomposite for tissue engineering applications. ACS Appl. Mater. Interfaces 2017, 9, 19356–19370.
- Yu, X.; Cheng, H.; Zhang, M.; Zhao, Y.; Qu, L.; Shi, G. Graphene-based smart materials. Nat. Rev. Mater. 2017, 2, 17046.
- Rolfe, P. Micro- and Nanosensors for Medical and Biological Measurement. Sens. Mater. 2012, 24, 275–302.
- Fang, Y. Label-free biosensors for cell biology. Int. J. Electrochem. 2011, 2011, 460850.
- Arroyo, N. Extending the Stability of Electrochemical, Aptamer-Based Sensors In Vivo. In ECS Meeting Abstracts; IOP Publishing: Bristol, UK, 2020; p. 3423.
- Chen, S.; Yuan, R.; Chai, Y.; Hu, F. Electrochemical sensing of hydrogen peroxide using metal nanoparticles: A review. Microchim. Acta 2013, 180, 15–32.
- Awan, M.; Rauf, S.; Abbas, A.; Nawaz, M.H.; Yang, C.; Shahid, S.A.; Amin, N.; Hayat, A. A sandwich electrochemical immunosensor based on antibody functionalized-silver nanoparticles (Ab-Ag NPs) for the detection of dengue biomarker protein NS1. J. Mol. Liq. 2020, 317, 114014.
- Power, A.C.; Gorey, B.; Chandra, S.; Chapman, J. Carbon nanomaterials and their application to electrochemical sensors: A review. Nanotechnol. Rev. 2018, 7, 19–41.
- Ibrahim, H.M.; Klingner, A. A review on electrospun polymeric nanofibers: Production parameters and potential applications. Polym. Test. 2020, 90, 106647.
- Long, Y.-Z.; Li, M.-M.; Gu, C.; Wan, M.; Duvail, J.-L.; Liu, Z.; Fan, Z. Recent advances in synthesis, physical properties and applications of conducting polymer nanotubes and nanofibers. Prog. Polym. Sci. 2011, 36, 1415–1442.
- Sapountzi, E.; Braiek, M.; Chateaux, J.-F.; Jaffrezic-Renault, N.; Lagarde, F. Recent advances in electrospun nanofiber interfaces for biosensing devices. Sensors 2017, 17, 1887.
- Zhang, M.; Zhao, X.; Zhang, G.; Wei, G.; Su, Z. Electrospinning design of functional nanostructures for biosensor applications. J. Mater. Chem. B 2017, 5, 1699–1711.
- Chen, Z.; Chen, Z.; Zhang, A.; Hu, J.; Wang, X.; Yang, Z. Electrospun nanofibers for cancer diagnosis and therapy. Biomater. Sci. 2016, 4, 922–932.
- Burugapalli, K.; Wijesuriya, S.; Wang, N.; Song, W. Biomimetic electrospun coatings increase the in vivo sensitivity of implantable glucose biosensors. J. Biomed. Mater. Res. Part A 2018, 106, 1072–1081.
- Saylan, Y.; Yilmaz, F.; Özgür, E.; Derazshamshir, A.; Yavuz, H.; Denizli, A. Molecular imprinting of macromolecules for sensor applications. Sensors 2017, 17, 898.
- Lv, Y.; Tan, T.; Svec, F. Molecular imprinting of proteins in polymers attached to the surface of nanomaterials for selective recognition of biomacromolecules. Biotechnol. Adv. 2013, 31, 1172–1186.
- Luo, J.; Cong, J.; Liu, J.; Gao, Y.; Liu, X. A facile approach for synthesizing molecularly imprinted graphene for ultrasensitive and selective electrochemical detecting 4-nitrophenol. Anal. Chim. Acta 2015, 864, 74–84.
- Anirudhan, T.; Deepa, J.; Stanly, N. Fabrication of a molecularly imprinted silylated graphene oxide polymer for sensing and quantification of creatinine in blood and urine samples. Appl. Surf. Sci. 2019, 466, 28–39.
- Uzun, L.; Turner, A.P. Molecularly-imprinted polymer sensors: Realising their potential. Biosens. Bioelectron. 2016, 76, 131–144.
- Ambrosi, A.; Pumera, M. Nanographite impurities dominate electrochemistry of carbon nanotubes. Chem. Eur. J. 2010, 16, 10946–10949.
More
Information
Subjects:
Biochemical Research Methods
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.0K
Revisions:
2 times
(View History)
Update Date:
13 Dec 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





