Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Krista G Freeman | + 2857 word(s) | 2857 | 2021-11-30 09:22:10 | | | |
| 2 | Rita Xu | Meta information modification | 2857 | 2021-12-06 02:54:52 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Freeman, K. A Mycobacteriophage-Based Vaccine Platform for SARS-CoV-2. Encyclopedia. Available online: https://encyclopedia.pub/entry/16728 (accessed on 07 February 2026).
Freeman K. A Mycobacteriophage-Based Vaccine Platform for SARS-CoV-2. Encyclopedia. Available at: https://encyclopedia.pub/entry/16728. Accessed February 07, 2026.
Freeman, Krista. "A Mycobacteriophage-Based Vaccine Platform for SARS-CoV-2" Encyclopedia, https://encyclopedia.pub/entry/16728 (accessed February 07, 2026).
Freeman, K. (2021, December 03). A Mycobacteriophage-Based Vaccine Platform for SARS-CoV-2. In Encyclopedia. https://encyclopedia.pub/entry/16728
Freeman, Krista. "A Mycobacteriophage-Based Vaccine Platform for SARS-CoV-2." Encyclopedia. Web. 03 December, 2021.
Copy Citation
Bacteriophage-based vaccines can generate a protective immune response by safely introducing foreign antigens displayed on, encapsidated within, or genetically encoded by phage. Here authors describe recombinants of mycobacteriophage Bxb1 (a phage infecting Mycobacterium smegmatis) that covalently display and express antigenic peptides of the SARS-CoV-2 Spike protein. Several of these vaccine candidates produced Spike-specific antibodies in immunized mice, but the responses were not neutralizing. This mycobacteriophage-based vaccine platform can likely be improved if delivery of larger antigens is achieved.
phage display vaccine
mycobacteriophage
SARS-CoV-2
DNA vaccine
1. Introduction
The COVID-19 pandemic illustrates the need for flexibility in vaccine development, and the potential roles of platforms such as those based on mRNA, adenovirus systems, and nanoparticles have been explored [1]. Continued exploration of phage-based systems offers the potential for new low cost, high production vaccines, and the global pandemic of SARS-CoV-2 provides a context for such development.
Bacteriophage vaccine platforms include the single-stranded DNA filamentous phage M13 platform used primarily in phage display technology [2][3][4][5] and several double-stranded DNA (dsDNA) tailed phage systems such as Escherichia coli phages T4 [6][7][8], T7 [9][10], and lambda [11][12][13][14]. Typically, the phage capsid of these dsDNA-tailed phages is used to display a foreign protein or part of a protein, most commonly through non-covalent linkages [4]. Several such recombinant phage systems have been shown to stimulate strong immune responses, which can be protective through neutralization of the infectious agent carrying the antigen [8][10][14]. Notably, two recent publications [15][16] describe innovative phage-based vaccine approaches for SARS-CoV-2, including phage display of antigens, delivery of phage genomes encoding antigens within mammalian expression cassettes, and encapsulation of antigenic proteins within the phage capsid. The evident advantages of such systems are the simplicity and relatively low cost of production, the adjuvantal nature of the phages themselves [17], and the strong safety profile of bacteriophages demonstrated during therapeutic use [18][19]. However, there are potential limitations with the need to prevent lipopolysaccharide (LPS) contamination from the E. coli host, growth and stability of the phages, and potential loss of antigen during purification [20]. Other phage systems are thus worth exploring, and a large number of phages of other bacterial hosts have been described.
The largest collection of phages known to infect a single common bacterial host are those of Mycobacterium smegmatis (referred to as mycobacteriophages). Over 10,000 have been isolated of which over 2000 genomes have been sequenced and annotated [21][22]. These are part of a larger collection of over 18,000 phages that infect various bacteria within the phylum Actinobacteria, over 3000 of which are genomically characterized [22]. Their genomes are highly diverse but can be grouped according to overall sequence relationships into ‘clusters’ (Cluster A, B, C, etc.), some of which can be readily divided into ‘subclusters’ (Subcluster A1, A2, A3, etc.) [22]. Some of the phages have no close relatives and these are referred to as ‘singletons’. The genetic diversity is such that even within these closely related clusters and subclusters there is substantial sequence variation and there are few examples of the same phage being independently isolated [22].
SARS-CoV-2 is a new coronavirus that has been intensely studied since its discovery in late 2019. Vaccine development efforts have focused on its spike (S) protein, which decorates the lipid membrane and makes direct contact with the human angiotensin-converting enzyme 2 (ACE2) receptor [23]. Vaccines developed by Moderna [24][25] and Pfizer [26] use mRNAs coding for the S protein, although with mutations that promote expression of the pre-fusion S protein conformation. The Oxford/AstraZeneca vaccine [27] delivers S protein DNA in an adenovirus vector, the Inovio vaccine [28] gives S DNA by electroporation, and the Novavax [29] platform uses nanoparticles decorated with S protein. Structural studies and comparisons with SARS-CoV-1 show that a 197-amino acid Receptor Binding Domain (RBD) is required for interaction with human ACE2, and that its 72-residue Receptor Binding Motif (RBM) makes direct contacts with ACE2 and is a target for neutralizing antibodies [30].
Mycobacteriophages have been used therapeutically to treat Mycobacterium infections and have strong safety profiles following intravenous administration [18]. Many grow well to high titer, are readily purified, and are stably maintained. Nonetheless, in immunocompetent patients extended intravenous application can elicit potent neutralizing antibody responses [31]. Understanding mycobacteriophage immunogenicity will therefore advance both their therapeutic use and their potential in vaccine development. No mycobacteriophage-based vaccines have been reported previously, although Mycobacterium-based vaccine platforms have been described, typically using mycobacteriophage-derived genetic tools [32][33].
2. Mycobacteriophage phiTM45 as a Vaccine Platform Candidate
All of the mycobacteriophages described to date are dsDNA-tailed phages with genomes ranging from 40–160 kbp. Most have isometric capsids with icosahedral symmetry, although there is considerable sequence variation among the morphogenic genes [22]. The capsids typically are constructed from pentameric and hexameric capsomers arranged with quasi-symmetry, and there are several examples of chainmail-like interlinking of the capsomers [34]. The tails are 130–350 nm long and composed of an abundant tail tube protein, a central tape measure protein, and an additional 6–10 minor tail proteins, including the receptor binding proteins.
We previously noted an unusual feature of mycobacteriophage Bxb1 virions (Figure 1A), in which both the capsid and the tail tube subunits contain C-terminal regions (~90 amino acids) that are sequence-related to each other but absent from closely related phages (Figure 1B) [35]. Because these C-terminal ‘extensions’ are absent from related phages they are presumably not essential for virion assembly. The similarity of the capsid protein to the well-studied HK97 suggests that the capsid extensions are present on the outside of the shell [36][37]. The specific role of these extensions is not known, but we note that this general relationship is seen with some other mycobacteriophages (e.g., Wildcat) where the extensions contain Ig-like motifs, as described for many other phages [38][39]. We reasoned that these protein segments could be replaced by antigens of choice, on either the capsid, tail tube, or both, and that dozens of different phages could be exploited in this way. A phage such as Bxb1 has a T = 7 symmetry, facilitating presentation of 415 copies of an antigen on a single capsid, and a tail of 130 nm length with ~270 copies of the tail tube protein [34]. We therefore explored whether these capsid and tail proteins could be exploited for antigen display.
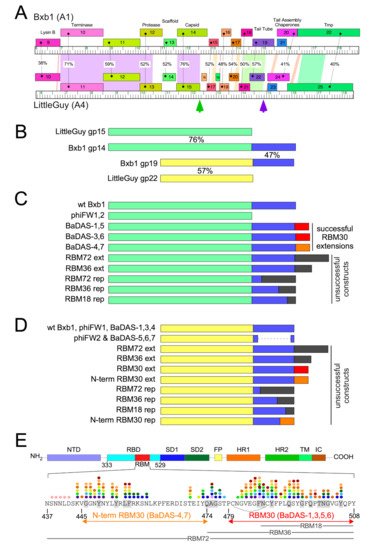
Figure 1. Design of Bacteriophage Displaying Antigens of SARS-CoV-2 (BaDAS) recombinant phages. (A) Alignment of phage Bxb1 (Subcluster A1) and LittleGuy (Cluster A4) virion structural genes. Genomes are represented as horizontal tracks with the predicted rightwards-transcribed genes shown as colored boxes above each genome. Gene names are shown in each box and genes are colored to indicate similarity. Spectrum colored shading between genomes shows pairwise BLASTN nucleotide alignment (violet strong similarity, red weakest similarity above threshold of BLASTN E-value of 10−4); amino acid identities (%) between genes are also shown. Green and purple vertical arrows indicate the Bxb1 capsid and tail tube C-terminal extensions. (B) Alignment of Bxb1 and LittleGuy capsid and tail tube proteins; shared capsid and tail tube protein segments are colored pale green and yellow, respectively, and the Bxb1 gp14 and gp19 extensions are shown in blue. Percent amino acid identities (%) are shown. (C) Schematic representations of Bxb1 recombinant phages. phiFW1 and phiFW2 lack the gp14 C-terminal extension, and BaDAS-1–BaDAS-7 have C-terminal SARS-CoV-2 RBM additions; red (RBM30) and orange (N-term RBM30) C-terminal additions correspond to the SARS-CoV-2 Spike protein regions in panel E. (D) Schematic representation of Bxb1 gp19 recombinant phages, colored as above. (E) SARS-CoV-2 Spike protein domain organization. Domains are depicted as described previously [23], with the Receptor Binding Domain (RBM) shown in red, and its sequence [40] shown below. The RBM segments displayed in BaDAS constructs (N-term RBM30 and RBM30, denoted with orange and red arrows, respectively) span residues binding to ACE2 [23][41][42] (gray highlighted residues) or various neutralizing antibodies, denoted with colored dots (see refs [43][44][45][46][47] for details). BaDAS-1, 3, 5, and 6 contain a disulfide bond stabilizing the RBM (indicated with an arc). The constructions containing regions RBM18, RBM36, and RBM72 (18, 36 and 72-residues, respectively, shown with gray labelled lines) were unsuccessful.
3. SARS-CoV-2 S Protein Epitopes for Display
We chose to display segments of the SARS-CoV-2 S RBM because it is both a critical component in viral infection as well as a vulnerable target for vaccines (Figure 1E). Several residues within the RBM form hydrogen bonds with the N-terminal helix of the human ACE2 [23][41][42], the receptor for SARS-CoV-2 infection [48][49]. Furthermore, many neutralizing antibodies against SARS-CoV-2 isolated from convalescent patient sera [43][44][45][46][47] bind to epitope residues within the RBM. The RBM is therefore a suitable component of SARS-CoV-2 to target for vaccine development. The ACE2-binding residues and neutralizing antibody epitope residues of the RBM are shown in Figure 1E. The segments chosen for constructing phage-based vaccine candidates span these critical residues.
4. Construction of the BaDAS Series of Vaccine Candidates
To engineer phage genomes we used the previously described CRISPY-BRED strategy [50]. In this approach, phage genomic DNA and a synthetic DNA substrate are electroporated into a recombineering strain of M. smegmatis to yield a mixture of parental and mutant phage progeny. These are then plated onto a strain expressing Cas9 and a single guide RNA (sgRNA) targeting the parental but not the mutant phage. The CRISPR-counter selection is typically sufficiently efficient that phage survivors are either the desired recombinants or CRISPR escape mutants (CEMs) with mutations in the targeted protospacer or associated PAM site [50]. The approach facilitates efficient construction of the desired recombinants, but also strongly indicates if a desired recombinant is non-viable, as then only CRISPR escape mutants are recovered among the progeny.
Using the CRISPY-BRED strategy we attempted to construct recombinant phages displaying antigenic peptides of SARS-CoV-2 on the surface of phiTM45, a lytic derivative of phage Bxb1 (Figure 1C,D and Figure 2A; Table 1); these are referred to as Bacteriophages Displaying Antigens of SARS-CoV-2 (BaDAS) vaccine candidates. We first demonstrated that the native C-terminal extensions on the capsid and tail tube proteins are nonessential by truncating them to form phages phiFW1 and phiFW2, respectively (Figure 1C,D). The sgRNA counter selection against parent phages is efficient (Figure 2B) and the constructions were uncomplicated, confirming that both derivatives are viable. Next, we attempted to replace the native C-terminal extensions with SARS-CoV-2 peptides of varying lengths (RBM72rep, RBM36rep, and RBM18rep, containing 72, 36, and 18 amino acids, respectively). Surprisingly, we were unable to recover any of these, and only CEMs were isolated among all the phage derivatives recovered and tested. Thus, although these extensions are not required for viability, they cannot be readily replaced by non-phage sequences in the way we had hoped. Nonetheless, when characterizing the CEMs that were recovered during attempted capsid engineering, we identified a mutant with a single base pair deletion in the protospacer that introduces a frameshift mutation near the 3′ end of the capsid gene, resulting in addition of 30 amino acids encoded in the -1 reading frame at the capsid protein C-terminus. This suggested that although the native capsid protein extension is dispensable—yet irreplaceable—that it could be extended by at least 30 residues. Based on this observation we made three recombinants (BaDAS-1, 3, and 4; for clarity, BaDAS-2 is the same as BaDAS-3, but contained a SNP and was not analyzed further) in which 30 amino acid segments of the SARS-CoV-2 RBM are added to the capsid C-terminus (Figures 1C and 2A,C); BaDAS-1 and BaDAS-3 have the RBM C-terminal residues added with slightly different junction sequences, and BaDAS-4 has the RBM N-terminal residues (Figure 1E). Similar constructions were attempted for the tail protein but were not able to be made (Figure 1D). However, the use of CRISPR to select against the 3′ end of the tail tube gene in this attempt yielded CEMs with truncated extensions of the tail tube, giving phages BaDAS-5, 6, and 7 (Figures 1C and 2D). The resulting six variants of genetically modified phages display > 400 antigenic peptides on the capsid surface, without evident interference of virion assembly or structure (as supported by viability). All phage derivatives were confirmed by Sanger sequencing in the region of the mutation; BaDAS-1 was completely sequenced.
Figure 2. Construction of BaDAS recombinant phages. (A) Modification of the Bxb1 capsid and tail tube subunits. Locations of the sgRNA (green line) targeting either the capsid or tail tube subunit genes (gene 14 and 19, top and bottom, respectively), and the synthetic DNA substrates (blue lines) carrying RBM sequences (red lines) are shown. (B) CRISPR-mediated counter selection. Plasmids with inducible expression of Cas9 and sgRNAs targeting the 3′ junctions of phage Bxb1 genes 14 or 19 (and its derivative phiTM45) were transformed into M. smegmatis mc2 155. Cells were plated with or without ATc inducer (as indicated) and 10-fold serial dilutions of Bxb1, phiTM45, and a control phage, BuzzLyseyear, were spotted onto the lawns. sgRNA induction reduces Bxb1 (and phiTM45) plaquing by at least four orders of magnitude. Top panel, targeting capsid gene 14; Bottom panel, targeting tail tube gene 19. (C) Primary plaques (left panel) recovered from CRISPY-BRED engineering were screened by PCR using primers flanking gene 14, and a candidate recombinant (candidate 3) was plaque purified and secondary plaques re-screened by PCR; all candidates are recombinant. (D) Primary plaques (left panel) recovered from CRISPR selection against wild type gene 19 were screened by PCR using primers flanking gene 19, and a candidate recombinant (candidate 5) was plaque purified and secondary plaques re-screened by PCR (right panel); all candidates are recombinant.
5. Construction of DEaDAS-1 Recombinant Vaccine Candidate
Bacteriophages can also be used to deliver DNA encoding targets for immune responses, as ‘DNA vaccines’ [51]. To explore whether these mycobacteriophages could be used similarly, we used the CRISPY-BRED approach to construct a recombinant carrying the S protein’s 197-residue RBD (Figure 1E) expressed from the cytomegalovirus (CMV) promoter (pCMV, Figure 3A). A 2495 bp mammalian expression cassette was amplified from pcDNA3-SARS-CoV-2-S-RBD-8his [52] (Addgene plasmid #145145, coordinates 138–2632) and ligated between two 250-bp gene blocks with homology to BaDAS-1, immediately upstream and downstream of gene 35 (Figure 3A). In a CRISPY-BRED experiment, the synthetic substrate and BaDAS-1 genomic DNA were co-electroporated into recombineering cells, and plaques were recovered on a sgRNA-expressing strain that counterselects against the parent phage (Figure 3B). Screening of primary plaques identified a desired recombinant (DNA Encoded and Displayed Antigens of SARS-CoV-2, DEaDAS) which was purified and confirmed by PCR and full genome sequencing (Figure 3C).
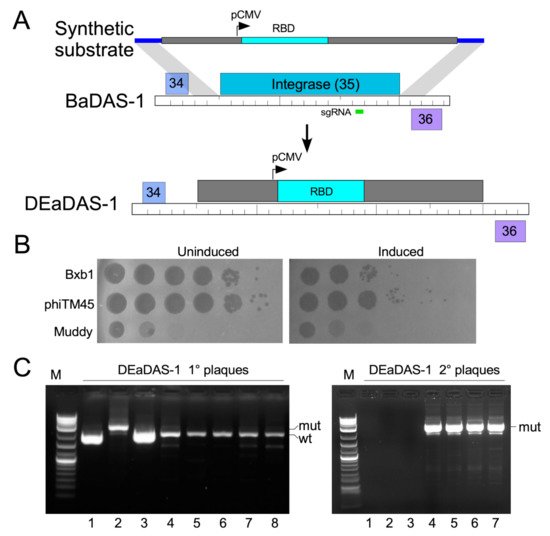
Figure 3. Construction of a DNA Encoded and Displayed Antigens of SARS-CoV-2 (DEaDAS) recombinant phage. (A) The integrase gene (blue) of phage phiTM45 was replaced with a mammalian expression cassette (gray box) containing the RBD of SARS-CoV2 (aqua) driven by a CMV promoter (pCMV). The location of the sgRNA target selecting against parent phiTM45 (green) is indicated. Regions of homology between the synthetic substrate and phiTM45 are indicated (blue line), and the structure of recombinant phage DEaDAS-1 is shown below. (B) sgRNA induction for selection against the integrase gene reduces Bxb1 and phiTM45 plaquing by at least two orders of magnitude. (C) Primary plaques (left panel) recovered from CRISPY-BRED engineering were screened by PCR, and a candidate recombinant (candidate 2) was plaque purified and secondary plaques (right panel) re-screened by PCR; all candidates are recombinant.
6. Characterization of Recombinant Phages
Recombination to make the desired phages seemingly occurred with relatively low frequency compared to other constructions [50]. This may be due to a minor growth defect, which is indicated by the difference in plaque sizes between the wild type and recombinant phages. While phiTM45, phiFW1, and phiFW2 all produce large plaques, BaDAS-1,-3, and -4 and DEaDAS-1 each form smaller plaques and produce raw lysates of slightly lower titer compared to phiTM45 (Figure 4A). Similarly, BaDAS-5,-6, and -7 exhibit plaque sizes slightly smaller than their full-length tail counterparts (Figure 4A).
Figure 4. Characterization of BaDAS and DEaDAS recombinant phages. (A) phiTM45 and derivatives phiFW1 and phiFW2 form large plaques on M. smegmatis, whereas all other derivatives form somewhat smaller plaques. (B) Thermostability of high titer, highly purified preparations of phiTM45 and vaccine candidates were assessed by determining the titer after incubation at ~4 °C for different times. All phages were stable for ~100 days of simple refrigeration. (C) SDS-PAGE of virion proteins shows the impact of modifications of capsid and tail tube subunits.
The choice of phage Bxb1 and its derivative phiTM45 as scaffolds for exploring vaccine construction was predicated in part because these can readily be grown to high titer and are stable when stored at 4 °C. To characterize the impact of virion modifications on phage stability, we incubated phiTM45, BaDAS-1, BaDAS-3, BaDAS-4, and DEaDAS-1 at ~ 4 °C and determined the phage titers at intervals up to at least 100 days (Figure 4B). All the phage preparations showed little or no reduction in titer over the 100-day period when stored cold at high (~1 × 1012 pfu/mL) concentration. These are encouraging parameters for potential vaccine development, especially in the context of a global pandemic where cold chain interruptions can spoil vaccines that must remain frozen.
Analysis of phage virion proteins confirmed the structures of the recombinant phages (Figure 4C). Phage phiTM45 has the expected profile with the most prominent bands being the tail tube subunit (predicted MW 30.1 kDa) and both pentameric and hexameric covalently interlinked capsid capsomers (predicted MW 209 and 250 kDa, respectively; Figure 4C) [34]. Phages phiFW2, BaDAS-5, BaDAS-6, and BaDAS-7 all have truncations of the tail tube gene and the major tail subunits are smaller as predicted, with molecular weights of ~23.5 kDa (Figure 4C). The truncation of the capsid subunit in phiFW1 and phiFW2 reduces the sizes of the capsid capsomers as expected (MW 164 and 197), and the small capsid additions in all BaDAS and DEaDAS phages have relatively little effect on capsid protein mobilities (Figure 4C).
References
- Haynes, B.F.; Corey, L.; Fernandes, P.; Gilbert, P.B.; Hotez, P.J.; Rao, S.; Santos, M.R.; Schuitemaker, H.; Watson, M.; Arvin, A. Prospects for a safe COVID-19 vaccine. Sci. Transl. Med. 2020, 12, eabe0948.
- Saw, P.E.; Song, E.W. Phage display screening of therapeutic peptide for cancer targeting and therapy. Protein Cell 2019, 10, 787–807.
- Shi, H.; Dong, S.; Zhang, X.; Chen, X.; Gao, X.; Wang, L. Phage vaccines displaying YGKDVKDLFDYAQE epitope induce protection against systemic candidiasis in mouse model. Vaccine 2018, 36, 5717–5724.
- Bao, Q.; Li, X.; Han, G.; Zhu, Y.; Mao, C.; Yang, M. Phage-based vaccines. Adv. Drug Deliv. Rev. 2019, 145, 40–56.
- Deng, L.; Ibanez, L.I.; Van den Bossche, V.; Roose, K.; Youssef, S.A.; de Bruin, A.; Fiers, W.; Saelens, X. Protection against Influenza A Virus Challenge with M2e-Displaying Filamentous Escherichia coli Phages. PLoS ONE 2015, 10, e0126650.
- Wu, J.; Tu, C.; Yu, X.; Zhang, M.; Zhang, N.; Zhao, M.; Nie, W.; Ren, Z. Bacteriophage T4 nanoparticle capsid surface SOC and HOC bipartite display with enhanced classical swine fever virus immunogenicity: A powerful immunological approach. J. Virol. Methods 2007, 139, 50–60.
- Ren, Z.J.; Lewis, G.K.; Wingfield, P.T.; Locke, E.G.; Steven, A.C.; Black, L.W. Phage display of intact domains at high copy number: A system based on SOC, the small outer capsid protein of bacteriophage T4. Protein Sci. 1996, 5, 1833–1843.
- Ren, Z.J.; Tian, C.J.; Zhu, Q.S.; Zhao, M.Y.; Xin, A.G.; Nie, W.X.; Ling, S.R.; Zhu, M.W.; Wu, J.Y.; Lan, H.Y.; et al. Orally delivered foot-and-mouth disease virus capsid protomer vaccine displayed on T4 bacteriophage surface: 100% protection from potency challenge in mice. Vaccine 2008, 26, 1471–1481.
- Wong, C.L.; Sieo, C.C.; Tan, W.S. Display of the VP1 epitope of foot-and-mouth disease virus on bacteriophage T7 and its application in diagnosis. J. Virol. Methods 2013, 193, 611–619.
- Wong, C.L.; Yong, C.Y.; Muhamad, A.; Syahir, A.; Omar, A.R.; Sieo, C.C.; Tan, W.S. A 12-residue epitope displayed on phage T7 reacts strongly with antibodies against foot-and-mouth disease virus. Appl. Microbiol. Biotechnol. 2018, 102, 4131–4142.
- Gonzalez-Cano, P.; Gamage, L.N.A.; Marciniuk, K.; Hayes, C.; Napper, S.; Hayes, S.; Griebel, P.J. Lambda display phage as a mucosal vaccine delivery vehicle for peptide antigens. Vaccine 2017, 35, 7256–7263.
- Hayes, S. Bacterial Virus Lambda Gpd-Fusions to Cathelicidins, alpha- and beta-Defensins, and Disease-Specific Epitopes Evaluated for Antimicrobial Toxicity and Ability to Support Phage Display. Viruses 2019, 11, 869.
- Hayes, S.; Gamage, L.N.; Hayes, C. Dual expression system for assembling phage lambda display particle (LDP) vaccine to porcine Circovirus 2 (PCV2). Vaccine 2010, 28, 6789–6799.
- Gamage, L.N.; Ellis, J.; Hayes, S. Immunogenicity of bacteriophage lambda particles displaying porcine Circovirus 2 (PCV2) capsid protein epitopes. Vaccine 2009, 27, 6595–6604.
- Staquicini, D.I.; Tang, F.H.F.; Markosian, C.; Yao, V.J.; Staquicini, F.I.; Dodero-Rojas, E.; Contessoto, V.G.; Davis, D.; O'Brien, P.; Habib, N.; et al. Design and proof of concept for targeted phage-based COVID-19 vaccination strategies with a streamlined cold-free supply chain. Proc. Natl. Acad. Sci. USA 2021, 118, e2105739118.
- Zhu, J.; Ananthaswamy, N.; Jain, S.; Batra, H.; Tang, W.C.; Lewry, D.A.; Richards, M.L.; David, S.A.; Kilgore, P.B.; Sha, J.; et al. A Universal Bacteriophage T4 Nanoparticle Platform to Design Multiplex SARS-CoV-2 Vaccine Candidates by CRISPR Engineering. bioRxiv 2021.
- Gonzalez-Mora, A.; Hernandez-Perez, J.; Iqbal, H.M.N.; Rito-Palomares, M.; Benavides, J. Bacteriophage-Based Vaccines: A Potent Approach for Antigen Delivery. Vaccines 2020, 8, 504.
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733.
- Schooley, R.T.; Biswas, B.; Gill, J.J.; Hernandez-Morales, A.; Lancaster, J.; Lessor, L.; Barr, J.J.; Reed, S.L.; Rohwer, F.; Benler, S.; et al. Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails To Treat a Patient with a Disseminated Resistant Acinetobacter baumannii Infection. Antimicrob. Agents Chemother. 2017, 61, e00954–17.
- Schooley, R.T.; Strathdee, S. Treat phage like living antibiotics. Nat. Microbiol. 2020, 5, 391–392.
- Russell, D.A.; Hatfull, G.F. PhagesDB: The actinobacteriophage database. Bioinformatics 2017, 33, 784–786.
- Hatfull, G.F. Actinobacteriophages: Genomics, Dynamics, and Applications. Annu. Rev. Virol. 2020, 7, 37–61.
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220.
- Corbett, K.S.; Edwards, D.K.; Leist, S.R.; Abiona, O.M.; Boyoglu-Barnum, S.; Gillespie, R.A.; Himansu, S.; Schafer, A.; Ziwawo, C.T.; DiPiazza, A.T.; et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 2020, 586, 567–571.
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931.
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Perez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615.
- Folegatti, P.M.; Ewer, K.J.; Aley, P.K.; Angus, B.; Becker, S.; Belij-Rammerstorfer, S.; Bellamy, D.; Bibi, S.; Bittaye, M.; Clutterbuck, E.A.; et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 2020, 396, 467–478.
- Tebas, P.; Yang, S.; Boyer, J.D.; Reuschel, E.L.; Patel, A.; Christensen-Quick, A.; Andrade, V.M.; Morrow, M.P.; Kraynyak, K.; Agnes, J.; et al. Safety and immunogenicity of INO-4800 DNA vaccine against SARS-CoV-2: A preliminary report of an open-label, Phase 1 clinical trial. EClinicalMedicine 2020, 31, 100689.
- Guebre-Xabier, M.; Patel, N.; Tian, J.H.; Zhou, B.; Maciejewski, S.; Lam, K.; Portnoff, A.D.; Massare, M.J.; Frieman, M.B.; Piedra, P.A.; et al. NVX-CoV2373 vaccine protects cynomolgus macaque upper and lower airways against SARS-CoV-2 challenge. Vaccine 2020, 38, 7892–7896.
- Liu, L.; Wang, P.; Nair, M.S.; Yu, J.; Rapp, M.; Wang, Q.; Luo, Y.; Chan, J.F.; Sahi, V.; Figueroa, A.; et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 2020, 584, 450–456.
- Dedrick, R.M.; Freeman, K.G.; Nguyen, J.A.; Bahadirli-Talbott, A.; Smith, B.E.; Wu, A.E.; Ong, A.S.; Lin, C.T.; Ruppel, L.C.; Parrish, N.M.; et al. Potent antibody-mediated neutralization limits bacteriophage treatment of a pulmonary Mycobacterium abscessus infection. Nat. Med. 2021, 27, 1357–1361.
- Broset, E.; Calvet Seral, J.; Arnal, C.; Uranga, S.; Kanno, A.I.; Leite, L.C.C.; Martin, C.; Gonzalo-Asensio, J. Engineering a new vaccine platform for heterologous antigen delivery in live-attenuated Mycobacterium tuberculosis. Comput. Struct. Biotechnol. J. 2021, 19, 4273–4283.
- Stover, C.K.; de la Cruz, V.F.; Fuerst, T.R.; Burlein, J.E.; Benson, L.A.; Bennett, L.T.; Bansal, G.P.; Young, J.F.; Lee, M.H.; Hatfull, G.F.; et al. New use of BCG for recombinant vaccines. Nature 1991, 351, 456–460.
- Mediavilla, J.; Kriakov, J.; Ford, M.E.; Duda, R.L.; Jacobs, W.R.; Hendrix, R.W.; Hatfull, G.F. Genome organization and characterization of mycobacteriophage Bxb1. Mol. Microbiol. 2000, 38, 955–970.
- Hatfull, G.F. Mycobacteriophages. In The Bacteriophages; Calendar, R., Ed.; Oxford University Press: New York, NY, USA, 2006; pp. 602–620.
- Duda, R.L. Protein Chainmail: Catenated Protein in Viral Capsids. Cell 1998, 94, 55–60.
- Wikoff, W.R.; Duda, R.L.; Hendrix, R.W.; Johnson, J.E. Crystallographic analysis of the dsDNA bacteriophage HK97 mature empty capsid. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999, 55, 763–771.
- Fraser, J.S.; Yu, Z.; Maxwell, K.L.; Davidson, A.R. Ig-like domains on bacteriophages: A tale of promiscuity and deceit. J. Mol. Biol. 2006, 359, 496–507.
- Campbell, P.L.; Duda, R.L.; Nassur, J.; Conway, J.F.; Huet, A. Mobile Loops and Electrostatic Interactions Maintain the Flexible Tail Tube of Bacteriophage Lambda. J. Mol. Biol. 2020, 432, 384–395.
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269.
- Yan, R.; Zhang, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448.
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224.
- Ju, B.; Zhang, Q.; Ge, J.; Wang, R.; Sun, J.; Ge, X.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020, 584, 115–119.
- Yi, C.; Sun, X.; Ye, J.; Ding, L.; Liu, M.; Yang, Z.; Lu, X.; Zhang, Y.; Ma, L.; Gu, W.; et al. Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell. Mol. Immunol. 2020, 17, 621–630.
- Shi, R.; Shan, C.; Duan, X.; Chen, Z.; Liu, P.; Song, J.; Song, T.; Bi, X.; Han, C.; Wu, L.; et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 2020, 584, 120–124.
- Kreye, J.; Reincke, S.M.; Kornau, H.C.; Sanchez-Sendin, E.; Corman, V.M.; Liu, H.; Yuan, M.; Wu, N.C.; Zhu, X.; Lee, C.D.; et al. A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model. Cell 2020, 183, 1058–1069.
- Barnes, C.O.; Jette, C.A.; Abernathy, M.E.; Dam, K.A.; Esswein, S.R.; Gristick, H.B.; Malyutin, A.G.; Sharaf, N.G.; Huey-Tubman, K.E.; Lee, Y.E.; et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 2020, 588, 682–687.
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569.
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273.
- Wetzel, K.S.; Guerrero-Bustamante, C.A.; Dedrick, R.M.; Ko, C.C.; Freeman, K.G.; Aull, H.G.; Divens, A.M.; Rock, J.M.; Zack, K.M.; Hatfull, G.F. Crispy-bred and crispy-brip: Efficient bacteriophage engineering. Sci. Rep. 2021, 11, 6796.
- Clark, J.R.; Bartley, K.; Jepson, C.D.; Craik, V.; March, J.B. Comparison of a bacteriophage-delivered DNA vaccine and a commercially available recombinant protein vaccine against hepatitis B. FEMS Immunol. Med. Microbiol. 2011, 61, 197–204.
- Chan, K.K.; Dorosky, D.; Sharma, P.; Abbasi, S.A.; Dye, J.M.; Kranz, D.M.; Herbert, A.S.; Procko, E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science 2020, 369, 6508.
More
Information
Subjects:
Microbiology; Virology; Immunology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
947
Entry Collection:
COVID-19
Revisions:
2 times
(View History)
Update Date:
06 Dec 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




