Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Katrin Voelter | + 1945 word(s) | 1945 | 2021-11-22 03:21:11 | | | |
| 2 | Rita Xu | -7 word(s) | 1938 | 2021-12-03 09:38:45 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Voelter, K. Recessive Achromatopsia in Original Braunvieh Cattle. Encyclopedia. Available online: https://encyclopedia.pub/entry/16713 (accessed on 07 February 2026).
Voelter K. Recessive Achromatopsia in Original Braunvieh Cattle. Encyclopedia. Available at: https://encyclopedia.pub/entry/16713. Accessed February 07, 2026.
Voelter, Katrin. "Recessive Achromatopsia in Original Braunvieh Cattle" Encyclopedia, https://encyclopedia.pub/entry/16713 (accessed February 07, 2026).
Voelter, K. (2021, December 03). Recessive Achromatopsia in Original Braunvieh Cattle. In Encyclopedia. https://encyclopedia.pub/entry/16713
Voelter, Katrin. "Recessive Achromatopsia in Original Braunvieh Cattle." Encyclopedia. Web. 03 December, 2021.
Copy Citation
Sporadic occurrence of inherited eye disorders has been reported in cattle but so far pathogenic variants were found only for rare forms of cataract but not for retinopathies.
Bos taurus
animal model
day-blindness
retina
development
mendelian genetics
1. Introduction
Food animal ophthalmology is a neglected area of veterinary medicine [1][2]. In farm animal practice, conditions such as visual impairment or even blindness, despite having a negative impact on behavior and welfare, are rarely considered, as there is usually no profoundly damaging economic impact on animal production [3].
Sporadic occurrence of inherited eye disorders in livestock species such as cattle has been reported [4][5][6]. However, many affected newborns go unreported or escape surveillance systems [4]. In addition to hereditary reasons, environmental causes such as vitamin A deficiency and bacterial or viral infections must also be taken into account [5][6][7]. Although congenital eye defects are rare, they are important and should be considered because they often follow monogenic recessive inheritance [4][6]. Therefore, such inherited disorders can rapidly gain in prevalence due to the undetected use of carriers, especially in the course of artificial insemination in cattle. So far pathogenic variants were found only for rare breed-specific recessive forms of cataract in Romagnola (OMIA 001936-9913) and Holstein (OMIA 002111-9913) cattle [8][9]. Such findings enable selection against these disorders within the affected populations. Furthermore, the NID1-related cataract observed in Romagnola was the first report of a naturally occurring mutation leading to a non-syndromic form of cataract in a mammalian species [8]. Therefore, the genetic study performed in cattle added the affected gene to the list of candidate genes for inherited forms of nuclear cataract in humans, illustrating the impact of studying eye conditions in domestic animals.
Achromatopsia, an inherited retinal disease characterized by the loss of cone photoreceptor function resulting in day-blindness, total color-blindness, and decreased central visual acuity, has not yet been described in cattle. Pathogenic variants in six genes (CNGA3, CNGB3, GNAT2, ATF6, PDE6C, and PDE6H) have been identified in humans with achromatopsia (OMIM 216900) [10]. In sheep, a form of CNGA3-related achromatopsia has been characterized (OMIA 001481-9940) [11]. This ovine condition was intensively used for functional restoration of cone function [12], highlighting the biomedical value of such large animal models [13][14] in addition to the direct benefits for animal breeding and animal health.
2. Clinical Description
Initially, in 2017, a four-month-old calf (case 2) was presented to the University of Zurich for evaluation of suspected day blindness. Ocular examination revealed bilaterally absent menace responses and dazzle reflexes. Pupillary responses were positive. Chromatic light stimulation (Melan) revealed bilaterally minimal dazzle reflexes with red and blue light, pupillary responses were normal with blue and reduced with red light stimulation. The remainder of the ocular exam (slit-lamp biomicroscopy, indirect and direct ophthalmoscopy, intraocular pressure) was within normal limits with the optic nerve head showing a deep physiologic cup. The calf was able to complete an obstacle course in dim light, but not in bright light.
In 2020, a 5.5-month-old calf (case 12) was presented to the University of Bern for evaluation of a suspected vision disturbance. The farmer noted that the calf was hesitant to walk and often collided with obstacles in its environment, especially when separated from the mother. Observation of the calf revealed extremely poor navigation of large and small objects in the examination hall in ambient room light. Navigation of obstacles and recognition of large high-contrast obstacles after room lights were turned off was slightly improved. Ophthalmic examination revealed absent menace response in both eyes (OU) in dim, ambient and bright (outdoor) light conditions. The direct and consensual pupillary light responses as well as dazzle reflexes to white light were reduced in both eyes. A slightly reduced pupillary light response was noted with bright blue light stimulation and severely reduced to bright red light stimulation. Slit-lamp biomicroscopic examination of the eyelids, conjunctiva, cornea, anterior chamber, lens and anterior vitreous was normal in both eyes with the exception of mild mydriasis OU. The pupils were symmetrical OU. Fluorescein staining of the cornea was negative OU. Indirect ophthalmoscopy revealed normal appearance to the optic nerve, tapetal and non-tapetal retina, and retinal vessels bilaterally. Intraocular pressure was measured within normal limits (11 mmHg OD, 14 mmHg OS).
Electroretinography in the affected calf revealed severely reduced light-adapted single cone and cone flicker responses, while the dark-adapted mixed cone-rod response was considered normal (Figure 1). Combined with the behavioral observations and clinical findings, the ERGs support an achromatopsia diagnosis based on the specific loss of cone-mediated retinal function without any retinal degenerative changes.
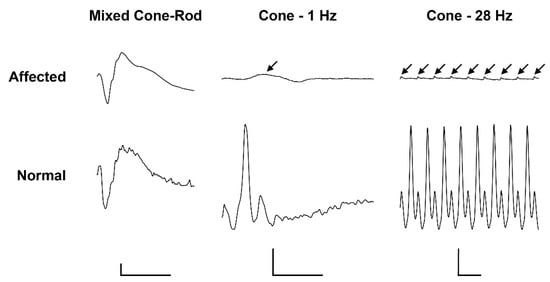
Figure 1. Electroretinogramm (ERG) of a 5.5-month old achromatopsia-affected Original Braunvieh (case 12) and a seven-month old Hereford control cattle. Dark-adaptation responses in representative normal and affected calves. While the dark-adapted mixed cone-rod responses were comparable between the two animals, the light-adapted single cone (1 Hz) and cone flicker (28 Hz) responses were severely reduced (arrows). Calibration bars: vertical = 100 µV, horizontal = 50 ms.
3. Pathological Phenotype
During necropsy of case 12 no gross lesions were detectable and brain areas involved in vision were morphologically normal.
The retina were targeted by immunofluorescence. We observed the expression of CNGB3 in the outer segment (OS) of cones in the control animal, while this expression was decreased in the affected animal. We also detected a significant difference of staining in the outer nuclear layer (ONL), where the CNGB3 protein seemed to be concentrated in the affected animal. This may suggest a mislocalization of the mutated CNGB3. Immunoreactivity of CNGB3 in the outer plexiform layer (OPL) may be due to an unspecific signal as the localization is observed in both control and affected animals. (Figure 2a). Further experiments on other affected calves (we analysed only one) or by transfection of the mutated CNGB3 in cells will confirm the potential higher stability of the mutated cyclic nucleotide-gated channel.
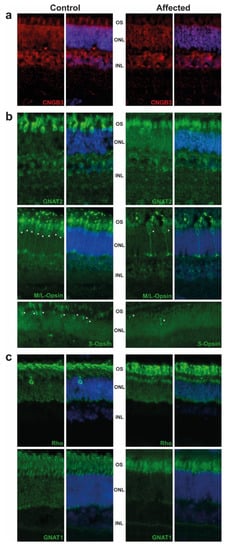
Figure 2. Immunostaining of retinal markers in a five-month old achromatopsia-affected cattle (case 12) and a control cattle of the same age. Cyclic nucleotide gated channel subunit beta 3 (CNGB3) (a), Cones markers (GNAT2, M/L-OPSIN, S-OPSIN) (b) and rods markers (GNAT1, RHODOPSIN) (c) are immunostained in both control and affected animals. Cell nuclei are shown in blue with DAPI. Images acquired at equal distances from the optic nerve head for each protein. Asterisks (*) show M/L- and S-Opsin cones present in both control and affected animal. Negative controls without primary antibody were performed (not shown).
Based on GNAT2 and cone opsins staining, we clearly observed a decrease in the cone OS, which are shorter and abnormally shaped. Although we did not count the number of cones per retina, we observed a decrease in their number, in affected animal in comparison to control, as shown in M/L-and S-Opsin staining (Figure 2b, white asterisk). These results are in correlation with the ERG and are consistent with the progressive loss of cone outer segments seen in other species with achromatopsia. We also observed a slight decrease of the rod OS length in affected animals and a mislocalization of Rho (Figure 2c).
4. Pedigree Analysis
The initially studied cases 1 and 2 were both the only affected animals in two different Swiss herds of purebred Original Braunvieh cattle. The sire of these two cases was a natural service purebred Original Braunvieh bull, which sired a further 24 apparently normal offspring within two years. A query to the Original Braunvieh breeders in Switzerland revealed further evidence of ten similar cases sired by different bulls collected over a period of three years. The available pedigree records of all 12 cases were analyzed and multiple inbreeding loops between the parents were found (Figure 3). We detected a single common ancestor occurring 8–11 generations ago. Due to the obvious history of inbreeding, a recessive inherited condition was considered. In light of the obvious consanguinity as well as the apparently unaffected parents, we hypothesize that the achromatopsia-affected calves might be explained by a recessively inherited variant. The founding mutation thus probably occurred many generations before the cases occurred. The causal variant was probably spread by the common ancestor, an artificial insemination bull born in 1961, as well as by some of his male descendants.
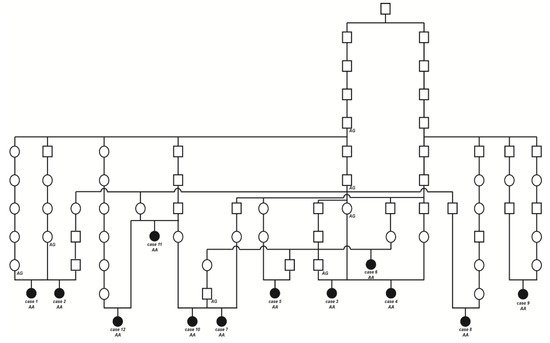
Figure 3. Pedigree of the Original Braunvieh cattle family segregating for achromatopsia suggested monogenic autosomal recessive inheritance. Affected animals are filled symbols. Open symbols represent normal cattle. DNA samples were available from animals with genotypes for the CNGB3 XM_015474554.2:c.751G>A variant are given below the symbols.
5. Genetic Analysis
SNP genotyping data for twelve affected cattle identified two shared ROHs between all cases on chromosome 11 and 14 (Figure 4a). On chromosome 11 all animals were homozygous for 101 SNP markers from 66,668,989 to 68,938,216 corresponding to a repeatedly detected strong selection signature of the Original Braunvieh breed encompassing a genome region with 24 protein-coding genes [15][16][17]. As no candidate gene for a retinopathy was contained in that region, we focused on the second ROH found on chromosome 14. All twelve affected cattle were homozygous at 28 SNP markers on chromosome 14 from 74,306,245 to 76,800,429, which allowed the identification of a single disease-associated IBD haplotype shared by all cases, limiting the critical region to 2,494,184 bp on chromosome 14 (Figure 4a,b). Interestingly, the bovine homolog of CNGB3, a gene that causes achromatopsia in other species, maps to that genome region at 76 Mb (Figure 4c).
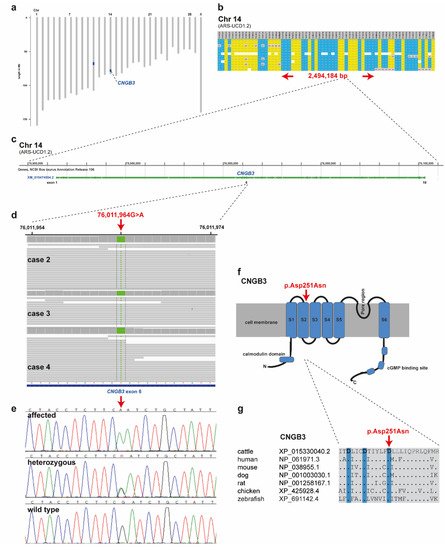
Figure 4. Achromatopsia-associated CNGB3 missense variant in Original Braunvieh. (a) Genome-wide homozygosity mapping presenting the two homozygous blocks shared in 12 affected calves in blue. Note the red arrow highlighting the CNGB3 gene on cattle chromosome 14. (b) Schematic representing the SNP genotypes of 12 affected calves on chromosome 14. Each horizontal lane represents one calf with yellow and blue shading, indicating shared homozygosity. Grey shading indicates a heterozygous genotype and white indicates missing genotypes. The genome positions of markers are indicated above the figure. The red arrows indicate the consensus homozygous region that spans approximately 2.5 Mb. (c) CNGB3 gene structure showing the location of the exon 6 variant. (d) Genome viewer screenshot presenting the homozygous Chr14: g.76011964A>G variant in three affected calves. (e) Electropherograms showing the different genotypes identified via Sanger sequencing. (f) Localization of the missense variant (red arrow) with respect to the topological model of the CNGB3 protein. (g) Across species sequence alignment of the affected CNGB3 S2 domain. Note that the missense variant (red arrow) affects the evolutionary conserved Tri-Asp motif that is highlighted in blue.
Subsequently, we sequenced the genomes of three of the affected cattle (cases 2, 3 and 4) and searched for private variants that were exclusively present in a homozygous state in all three affected cattle and absent or only heterozygous in the genomes of 567 other cattle. Beside 34 non-coding variants, all located in the critical region on chromosome 14, this analysis identified a single homozygous private protein-changing variant in CNGB3, a known candidate gene for achromatopsia. The variant can be designated as chr14: 76011964G>A (ARS-UCD1.2 assembly) (Figure 4d). It is a missense variant, XM_015474554.2: c.751G>A, predicted to change a highly conserved aspartic acid residue in the second S2 domain of CNGB3, XP_015330040.2: p.Asp251Asn (Figure 4f,g). In silico analysis predicted the functional effect of p.Asp251Asn as deleterious using PROVEAN software (score −4.985) [18].
Authors confirmed the presence of the CNGB3 missense variant by Sanger sequencing (Figure 4e). The genotypes at the variant co-segregated with the achromatopsia phenotype as expected for a monogenic autosomal recessive mode of inheritance (Figure 3). All twelve available DNA samples from the achromatopsia-affected cattle carried the mutant allele in a homozygous state, while their parents were heterozygous, as expected for obligate carriers (Figure 3; Table 1).
Table 1. Association of the missense variant in CNGB3 with the achromatopsia phenotype in Original Braunvieh cattle.
| GG | AG | AA | |
|---|---|---|---|
| Achromatopsia-affected calves | 12 | ||
| Obligate carriers a | 5 | ||
| Other Original Braunvieh cattle b,c | 2477 | 463 | 12 |
| Brown Swiss cattle c | 14,976 | 52 | |
| Holstein cattle c | 14,825 | ||
| Simmental cattle c | 2021 | ||
| Sequenced cattle genomes from various breeds (local Swiss cohort) d | 552 | 15 | |
| Control cattle from various breeds (1000 Bull Genomes project) e | 3298 | 7 f | 1 g |
References
- Lavach, J.D. Large Animal Ophthalmology; Mosby: St. Louis, MO, USA, 1990.
- Williams, D.L. Production animal ophthalmology. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, xi–xii.
- Williams, D.L. Welfare issues in farm animal ophthalmology. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 427–435.
- Leipold, H.W. Congenital ocular defects in food-producing animals. Vet. Clin. N. Am. Large Anim. Pract. 1984, 6, 577–595.
- Gelatt, K.N.; Ben-Shlomo, G.; Gilger, B.C.; Hendrix, D.V.; Kern, T.J.; Plummer, C.E. Veterinary Ophthalmology, 6th ed.; John Wiley & Sons: Hoboken, NJ, USA, 2021; pp. 1983–2054.
- Williams, D.L. Congenital abnormalities in production animals. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 477–486.
- He, X.; Li, Y.; Li, M.; Jia, G.; Dong, H.; Zhang, Y.; He, C.; Wang, C.; Deng, L.; Yang, Y. Hypovitaminosis A coupled to secondary bacterial infection in beef cattle. BMC Vet. Res. 2012, 8, 222.
- Murgiano, L.; Jagannathan, V.; Calderoni, V.; Joechler, M.; Gentile, A.; Drögemüller, C. Looking the cow in the eye: Deletion in the NID1 gene is associated with recessive inherited cataract in Romagnola cattle. PLoS ONE 2014, 9, e110628.
- Hollmann, A.K.; Dammann, I.; Wemheuer, W.M.; Wemheuer, W.E.; Chilla, A.; Tipold, A.; Schulz-Schaeffer, W.J.; Beck, J.; Schütz, E.; Brenig, B. Morgagnian cataract resulting from a naturally occurring nonsense mutation elucidates a role of CPAMD8 in mammalian lens development. PLoS ONE 2017, 12, e0180665.
- Sun, W.; Li, S.; Xiao, X.; Wang, P.; Zhang, Q. Genotypes and phenotypes of genes associated with achromatopsia: A reference for clinical genetic testing. Mol. Vis. 2020, 26, 588–602.
- Reicher, S.; Seroussi, E.; Gootwine, E. A mutation in gene CNGA3 is associated with day blindness in sheep. Genomics 2010, 95, 101–104.
- Ross, M.; Ofri, R.; Aizenberg, I.; Abu-Siam, M.; Pe’er, O.; Arad, D.; Rosov, A.; Gootwine, E.; Dvir, H.; Honig, H.; et al. Naturally-occurring myopia and loss of cone function in a sheep model of achromatopsia. Sci. Rep. 2020, 10, 19314.
- Komáromy, A.M. Day blind sheep and the importance of large animal disease models. Vet. J. 2010, 185, 241–242.
- Winkler, P.A.; Occelli, L.M.; Petersen-Jones, S.M. Large animal models of inherited retinal degenerations: A review. Cells 2020, 9, 882.
- Rothammer, S.; Seichter, D.; Förster, M.; Medugorac, I. A genome-wide scan for signatures of differential artificial selection in ten cattle breeds. BMC Genom. 2013, 14, 908.
- Signer-Hasler, H.; Burren, A.; Neuditschko, M.; Frischknecht, M.; Garrick, D.; Stricker, C.; Gredler, B.; Bapst, B.; Flury, C. Population structure and genomic inbreeding in nine Swiss dairy cattle populations. Genet. Sel. Evol. 2017, 49, 83.
- Bhati, M.; Kadri, N.K.; Crysnanto, D.; Pausch, H. Assessing genomic diversity and signatures of selection in Original Braunvieh cattle using whole-genome sequencing data. BMC Genom. 2020, 21, 27.
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747.
More
Information
Subjects:
Health Care Sciences & Services
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
856
Revisions:
2 times
(View History)
Update Date:
03 Dec 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




