Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jinpeng Zou | + 1886 word(s) | 1886 | 2021-12-03 03:34:42 | | | |
| 2 | Camila Xu | + 1884 word(s) | 1884 | 2021-12-03 03:59:56 | | | | |
| 3 | Camila Xu | Meta information modification | 1884 | 2021-12-03 04:17:28 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Zou, J. Arabidopsis LSH8 ABA Signaling. Encyclopedia. Available online: https://encyclopedia.pub/entry/16705 (accessed on 08 February 2026).
Zou J. Arabidopsis LSH8 ABA Signaling. Encyclopedia. Available at: https://encyclopedia.pub/entry/16705. Accessed February 08, 2026.
Zou, Jinpeng. "Arabidopsis LSH8 ABA Signaling" Encyclopedia, https://encyclopedia.pub/entry/16705 (accessed February 08, 2026).
Zou, J. (2021, December 03). Arabidopsis LSH8 ABA Signaling. In Encyclopedia. https://encyclopedia.pub/entry/16705
Zou, Jinpeng. "Arabidopsis LSH8 ABA Signaling." Encyclopedia. Web. 03 December, 2021.
Copy Citation
LSH is a family of transcription factors with diversified functions, the members of which, in turn, are named LSH1-LSH10. LIGHT-DEPENDENT SHORT HYPOCOTYLS 1 (LSH1 was first found in Arabidopsis in 2004, the overexpression of which can enhance the light response of Arabidopsis thaliana seedlings and show an obvious short hypocotyl phenotype. All members of the LSH family have a highly conserved Domain of Unknown Function 640 (DUF640) domain, which is also called the Arabidopsis LSH1 and Oryza G1 (ALOG) domain in the Pfam protein database.
Arabidopsis
LSH8
ABA
seed germination
proteomics
ARPs
1. Introduction
As a sessile organism, plants need to undergo a complex internal regulation mechanism and environmental signal regulation to survive in adverse and changeable environments [1]. Phytohormone ABA is an important signaling regulator that plays a crucial role in mediating seed germination and maturation, seedling growth, stomatal movement, flowering and stress responses [1][2]. For example, ABA can regulate seed dormancy to prevent premature germination of seeds under stress conditions so that the seeds are able to germinate under suitable conditions, improving the germination rate [3]. These important functions of ABA are derived from the sophisticated regulatory network of ABA [4].
Current research demonstrates that the ABA signaling network in Arabidopsis includes five important components: ABA receptors with PYR1-like (PYL) components, negative regulator type 2C protein phosphatases (PP2C), positive regulator SNF1-related protein kinase 2 (SnRK2), transcription factors of basic leucine zippers (bZIP) and ABA-responsive genes [5]. The signal transduction of ABA in plants occurs in the following pathways. When ABA is deficient, PP2C with phosphatase activity dephosphorylates SnRK2 to inhibit the expression of downstream ABA-responsive genes activated by SnRK2, while in the presence of ABA, the complex of ABA binding to PYR/PYL/RCAR receptors inhibits the phosphatase activity of PP2C, from which SnRK2 is released. The released SnRK2 phosphorylates the downstream transcription factors ABI3/ABI4/ABI5 and ABA-response element binding factors (ABFs), thereby activating the expression of ABA-responsive genes [4][6][7][8][9][10][11]. Numerous previous studies have shown that a large number of transcription factors in the ABA signaling pathway play an indispensable role. For example, ABI3 is a B3-type transcription factor, and ABI5 is a bZIP transcription factor, both of which mediate ABA-induced inhibition of seed germination and initial seedling growth to participate in ABA signal transduction at the seedling stage [12][13][14]. Their loss-of-function mutation leads to the weakening of the inhibitory effect of ABA on seed germination [13][15]. Exogenous ABA significantly induces the expression of ABI5, the overexpression lines of which show a hypersensitivity phenotype to ABA during seed germination and early seedling development [16]. Furthermore, ABI5, as a bZIP transcription factor, can bind to the ABA binding response element (ABRE) in the promoter region of the target genes to activate the target genes’ expression [17]. Additionally, ABF1, ABF2/AREB1, ABF3, ABF4/AREB2 and other transcription factors have been reported to play important roles in the ABA signaling network [18][19], and after the phosphorylation caused by activated SnRK2, ABF/AREB directly binds to the promoter of stress response genes (such as RD29A and RD29B) to stimulate its transcriptional activity under stress conditions [19][20][21]. These reports suggest that a large number of transcription factors play an important role in the highly complex signaling network of ABA.
LSH is a family of transcription factors with diversified functions, the members of which, in turn, are named LSH1-LSH10. LIGHT-DEPENDENT SHORT HYPOCOTYLS 1 (LSH1 was first found in Arabidopsis in 2004 [22], the overexpression of which can enhance the light response of Arabidopsis thaliana seedlings and show an obvious short hypocotyl phenotype. All members of the LSH family have a highly conserved Domain of Unknown Function 640 (DUF640) domain, which is also called the Arabidopsis LSH1 and Oryza G1 (ALOG) domain in the Pfam protein database [22][23]. The DUF640/ALOG domain contains four all-α helices, the additional insertion of a zinc ribbon and a nuclear location signal (NLS) [24]. Proteins with the DUF640/ALOG domain comprise a class of specific transcription factors in plants, with characteristics of binding DNA sequence specificity, transcriptional regulation activity, nuclear localization and homodimer formation, and control plant growth and development in many aspects. Therefore, transcription factor proteins with such a domain often have specific functions [23][25][26][27]. Studies have found that LSH1 inhibits hypocotyl length in Arabidopsis thaliana in a light-dependent manner. The expression of LSH3 and LSH4 in the cells of various lateral organs, such as the cotyledon, the leaf and the flower organ, inhibits the differentiation of the boundary organ [25][28][29]. LSH9 interacts with the temperature sensor ELF3 to regulate hypocotyl elongation [30][31]. In addition, proteins of the LSH family can regulate inflorescence structure and flower organ development in other plant species [32]. LSH family genes extensively participate in different biological processes in plants, but whether its family genes participate in plant stress response remains unknown.
2. LSH8 Regulates Seed Germination and the Elongation of Primary and Lateral Root
LSH family genes are reported to be expressed in hypocotyl and flower organs. They are important for the growth and development of plants, but their function in the hormone signaling network remains unknown. To understand the function of LSH8 in the ABA signaling pathway, we obtained 35S::LUC-LSH8 overexpression lines LSH8-#2 and LSH8-#5, lsh8 mutant lines lsh8-1 (SALK_024841) and lsh8-2 (CS845710). However, ABA inhibition on the seed germination of lsh8 mutant lines lsh8-1 and lsh8-2 were obviously attenuated. Thus, LSH8 overexpression lines were recognized as ABA sensitive and lsh8 mutant lines insensitive (Figure 1A). As the seed germination is under the joint regulation of the hormones ABA and GA, during which GA promotes seed germination, presenting the opposite effect of ABA, we used GA biosynthesis inhibitor paclobutrazol (PAC) to verify the LSH8 phenotype and found that under the PAC treatment condition, the seed germination of different genotypes was identical to that under ABA treatment. At the same time, we applied an appropriate amount of GA on ABA treatment conditions, the result of which showed that GA could weaken the inhibitory effect of ABA on seed germination (Figure 1A). To summarize, the above results indicate that ABA and GA simultaneously participate in the process of seed germination. More importantly, we identified a new positive regulator LSH8 in the ABA signaling pathway. Further statistical results of germination rate showed that under either ABA or PAC treatment, the germination rate of lsh8 mutant lines was significantly higher than that of Col4, while the germination rate of LSH8 overexpression lines was significantly lower. Additionally, under ABA treatment with a moderate amount of GA, the seed germination rate of LSH8 overexpression lines and lsh8 mutant lines was improved (Figure 1B).
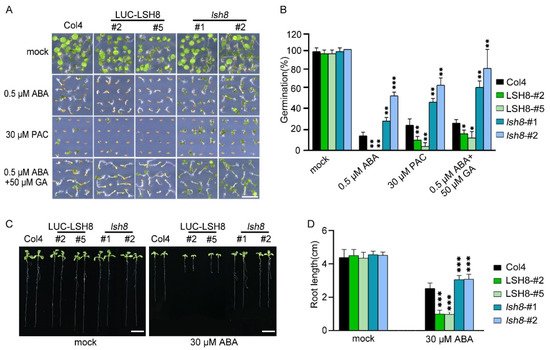
Figure 1. ABA phenotype of LSH8 overexpression and lsh8 mutant lines. (A) Germination phenotypes of LSH8 overexpression lines (LSH8-#2 and LSH8-#5), lsh8 mutant lines (lsh8-1 and lsh8-2) and wild type (Col4). Seeds were germinated and grown on 1/2 MS (mock) and 1/2 MS containing 0.5 µM ABA, 30 µM PAC, 0.5 µM ABA + 50 µM GA for 5 d, respectively. Scale bar: 1 cm. (B) Statistical analysis of germination rate described in (A). Data represent mean ± SD of at least 64 seeds. (C) Comparison of root length among genotypes on 1/2 MS with or without 30 µM ABA, respectively. Scale bar: 1 cm. (D) Statistical analysis of the differences in root length among the genotypes shown in (C). Data are shown as mean ± SD (n > 10). Asterisks in (B,D) indicate statistically significant differences compared with wild-type Col4: *, p < 0.05; **, p < 0.01; ***, p < 0.001 (Student’s t-test).
Since the LSH8-mediated ABA signaling pathway participates in the regulation of seed germination, whether LSH8 is involved in ABA-mediated seedling root elongation is another issue of concern. After growing on 1/2 MS medium without ABA for 4 days, the seeds of different genotypes were transferred to 1/2 MS medium with ABA for another 4 days. The root length phenotype and root length measurements showed that when compared with wild-type Col4, the primary roots of the overexpression lines LSH8-#2 and LSH8-#5 were significantly shorter, but the primary roots of mutant lines lsh8-1 and lsh8-2 were significantly longer, and their lateral roots were obviously increasing (Figure 1C,D), showing that LSH8 promotes ABA’s effect on inhibiting the primary root elongation and lateral root development.
3. The Prediction of Upstream Element of LSH8 and the Expression of LSH8 Regulated by ABA
In the ABA signaling pathway, ABREs can be recognized by specific transcription factors to activate the expression of ABA downstream related response genes. Most of the promoter region of ABA response genes contains conserved G-box-like cis-elements, ABREs (PyACGTGG/TC). Previous studies have shown that genes successfully activated and expressed by ABA require multiple ABREs or one ABRE bound to several coupling elements (CEs). Analyzing the data of PlantCare, we found that the promoter region of LSH8 contained two ABREs and one G-box element.
With ePlant (http://bar.utoronto.ca/eplant (accessed on 25 August 2020)), we predicted that LSH8 was localized in the nucleus. Further protoplast subcellular localization experiment showed that LSH8 was localized in the nucleus (Figure 2A). Additionally, to analyze the LSH8 expression in different tissues, we detected the root, the stem, the leaf, the flower and the silique of Arabidopsis by qPCR, the result of which showed that the highest expression of LSH8 happened in the flower and the silique and the lowest in the stem (Figure 2B). Previous studies have found that PhLSH7b of petunia, a homologous gene of LSH8 in Arabidopsis, regulates plant flowering, but the function of LSH8 in Arabidopsis is still unknown. To study the function of LSH8, we checked the public microarray data (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi (accessed on 13 September 2020)) and found that the expression of LSH8 may be affected by ABA during seed germination. To confirm this expression pattern, we detected the changes in the transcriptional level and protein level of LSH8 in wild-type Col4 under ABA treatment. The results showed that the transcriptional level of LSH8 started to have a gradual decrease after 1.5 h of ABA treatment (Figure 2C). With additional Luciferase assay and Western blot detection, we found that the protein level of LSH8 from LSH8 overexpression lines also showed a downtrend under ABA treatment (Figure 2D,E). The above results indicated that ABA inhibited the expression of LSH8 at both transcriptional and protein levels.
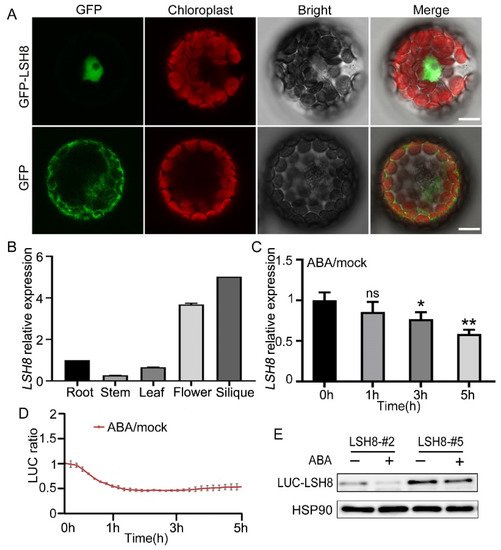
Figure 2. Expression pattern analysis of LSH8. (A) Subcellular localization analysis of GFP and GFP-LSH8 in Arabidopsis wild-type Col4 protoplasts. The channels from left to right are GFP, Chloroplast, Bright and Merged channels, respectively. Scale bar: 10 µm. (B) Tissue-specific expression of LSH8. Various tissues of wild-type Col4 were grown under normal conditions, and the LSH8 expression was determined by qPCR. Data are shown as mean ± SD (n = 3). (C) qPCR analysis of LSH8 transcriptional level under ABA treatment. Five-day-old Col4 seedlings were treated with 100 µM ABA for 0–5 h. Data are shown as mean ± SD (n = 3). (D) LUC signals in 5-d-old LUC-LSH8 overexpressing seedlings treated with 100 µM ABA. Signals were detected every 10 min, and the detecting period is 5 h. Data are shown as mean ± SD (n = 3). (E) Immunoblot analyzing the ABA-induced decline of LSH8 protein in the LUC-LSH8 overexpressing lines. Whole seedlings of 5-day-old LUC-LSH8 overexpression lines were treated with 100 µM ABA for 5 h. The expression of LUC-LSH8 fusion protein was detected by immunoblotting with an anti-LUC antibody. HSP90 was used as loading control. Asterisks in (C) indicate statistically significant differences compared with normal conditions (0 h): ns, p > 0.05; *, p < 0.05; **, p < 0.01 (Student’s t-test).
References
- Hauser, F.; Waadt, R.; Schroeder, J.I. Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 2011, 21, R346–R355. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Gampala, S.S.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14, S15–S45. [Google Scholar] [CrossRef]
- Verslues, P.E.; Zhu, J.K. New developments in abscisic acid perception and metabolism. Curr. Opin. Plant Biol. 2007, 10, 447–452. [Google Scholar] [CrossRef]
- Finkelstein, R. Abscisic Acid synthesis and response. Arab. Book 2013, 11, e0166. [Google Scholar] [CrossRef]
- Yang, W.; Zhang, W.; Wang, X. Post-translational control of ABA signalling: The roles of protein phosphorylation and ubiquitination. Plant Biotechnol. J. 2017, 15, 4–14. [Google Scholar] [CrossRef]
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Ishihama, Y.; Hirayama, T.; Shinozaki, K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593. [Google Scholar] [CrossRef]
- Vlad, F.; Rubio, S.; Rodrigues, A.; Sirichandra, C.; Belin, C.; Robert, N.; Leung, J.; Rodriguez, P.L.; Laurière, C.; Merlot, S. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 2009, 21, 3170–3184. [Google Scholar] [CrossRef] [PubMed]
- Bhaskara, G.B.; Nguyen, T.T.; Verslues, P.E. Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiol 2012, 160, 379–395. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant 2013, 147, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Furihata, T.; Maruyama, K.; Fujita, Y.; Umezawa, T.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 2006, 103, 1988–1993. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Giraudat, J.; Hauge, B.M.; Valon, C.; Smalle, J.; Parcy, F.; Goodman, H.M. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 1992, 4, 1251–1261. [Google Scholar]
- Finkelstein, R.R.; Lynch, T.J. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 2000, 12, 599–609. [Google Scholar] [CrossRef]
- Söderman, E.M.; Brocard, I.M.; Lynch, T.J.; Finkelstein, R.R. Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol. 2000, 124, 1752–1765. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Xue, Q.; McCray, T.; Margavage, K.; Chen, F.; Lee, J.H.; Nezames, C.D.; Guo, L.; Terzaghi, W.; Wan, J.; et al. The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell 2013, 25, 517–534. [Google Scholar] [CrossRef]
- Lopez-Molina, L.; Mongrand, S.; Chua, N.H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 2001, 98, 4782–4787. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Zhang, P.; Ho, T.H. Modular nature of abscisic acid (ABA) response complexes: Composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell 1996, 8, 1107–1119. [Google Scholar] [PubMed]
- Joshi, R.; Wani, S.H.; Singh, B.; Bohra, A.; Dar, Z.A.; Lone, A.A.; Pareek, A.; Singla-Pareek, S.L. Transcription Factors and Plants Response to Drought Stress: Current Understanding and Future Directions. Front. Plant Sci. 2016, 7, 1029. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. Cell Mol. Biol. 2010, 61, 672–685. [Google Scholar] [CrossRef] [PubMed]
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 2000, 97, 11632–11637. [Google Scholar] [CrossRef] [PubMed]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. Cell Mol. Biol. 2003, 34, 137–148. [Google Scholar] [CrossRef]
- Zhao, L.; Nakazawa, M.; Takase, T.; Manabe, K.; Kobayashi, M.; Seki, M.; Shinozaki, K.; Matsui, M. Overexpression of LSH1, a member of an uncharacterised gene family, causes enhanced light regulation of seedling development. Plant J. Cell Mol. Biol. 2004, 37, 694–706. [Google Scholar] [CrossRef]
- Yoshida, A.; Suzaki, T.; Tanaka, W.; Hirano, H.Y. The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proc. Natl. Acad. Sci. USA 2009, 106, 20103–20108. [Google Scholar] [CrossRef]
- Chen, F.; Zhou, Q.; Wu, L.; Li, F.; Liu, B.; Zhang, S.; Zhang, J.; Bao, M.; Liu, G. Genome-wide identification and characterization of the ALOG gene family in Petunia. BMC Plant Biol. 2019, 19, 600. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Hanano, K.; Kariya, A.; Shimizu, S.; Zhao, L.; Matsui, M.; Tasaka, M.; Aida, M. CUP-SHAPED COTYLEDON1 transcription factor activates the expression of LSH4 and LSH3, two members of the ALOG gene family, in shoot organ boundary cells. Plant J. Cell Mol. Biol. 2011, 66, 1066–1077. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Aravind, L. ALOG domains: Provenance of plant homeotic and developmental regulators from the DNA-binding domain of a novel class of DIRS1-type retroposons. Biol. Direct 2012, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Peng, P.; Liu, L.; Fang, J.; Zhao, J.; Yuan, S.; Li, X. The rice TRIANGULAR HULL1 protein acts as a transcriptional repressor in regulating lateral development of spikelet. Sci. Rep. 2017, 7, 13712. [Google Scholar] [CrossRef]
- Hibara, K.; Takada, S.; Tasaka, M. CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation. Plant J. Cell Mol. Biol. 2003, 36, 687–696. [Google Scholar] [CrossRef]
- Kamiuchi, Y.; Yamamoto, K.; Furutani, M.; Tasaka, M.; Aida, M. The CUC1 and CUC2 genes promote carpel margin meristem formation during Arabidopsis gynoecium development. Front. Plant Sci. 2014, 5, 165. [Google Scholar] [CrossRef]
- Press, M.O.; Queitsch, C. Variability in a Short Tandem Repeat Mediates Complex Epistatic Interactions in Arabidopsis thaliana. Genetics 2017, 205, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Hayes, S.; Schachtschabel, J.; Mishkind, M.; Munnik, T.; Arisz, S.A. Hot topic: Thermosensing in plants. Plant Cell Environ. 2021, 44, 2018–2033. [Google Scholar] [CrossRef]
- Yoshida, A.; Sasao, M.; Yasuno, N.; Takagi, K.; Daimon, Y.; Chen, R.; Yamazaki, R.; Tokunaga, H.; Kitaguchi, Y.; Sato, Y.; et al. TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc. Natl. Acad. Sci. USA 2013, 110, 767–772. [Google Scholar] [CrossRef] [PubMed]
More
Information
Subjects:
Cell Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
948
Revisions:
3 times
(View History)
Update Date:
03 Dec 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




