| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Tiago Santos | + 1679 word(s) | 1679 | 2021-08-12 09:47:15 | | | |
| 2 | Amina Yu | + 6 word(s) | 1685 | 2021-12-03 06:47:47 | | | | |
| 3 | Amina Yu | Meta information modification | 1685 | 2021-12-03 09:37:04 | | |
Video Upload Options
Progress in the design of G-quadruplex (G4) binding ligands relies on the availability of approaches that assess the binding mode and nature of the interactions between G4 forming sequences and their putative ligands. The experimental approaches used to characterize G4/ligand interactions can be categorized in structure-based methods (circular dichroism (CD), nuclear magnetic resonance (NMR) spectroscopy and x-ray crystallography), affinity and apparent affinity-based methods (surface plasmon resonance (SPR), isothermal titration calorimetry (ITC) and mass spectrometry (MS)), and high-throughput methods (fluorescence resonance energy transfer (FRET)-melting, G4-fluorescent intercalator displacement assay (G4-FID), affinity chromatography and microarrays. Each method has unique advantages and drawbacks, which makes it essential to select the ideal strategies for the biological question being addressed. The structural- and affinity and apparent affinity-based methods are in several cases complex and/or time-consuming and can be combined with fast and cheap high-throughput approaches to improve the design and development of new potential G4 ligands. In the last years, the joint use of these techniques permitted the discovery of a huge number of G4 ligands investigated for diagnostic and therapeutic purposes. Overall, this review article highlights in detail the most used approaches to characterize the G4/ligand interactions, as well as the applications and type of information that can be obtained from the use of each technique.
1. Introduction
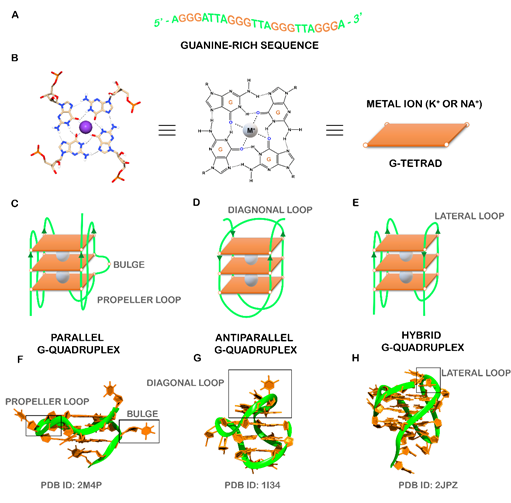
The human genome and transcriptome contain several guanine-rich sequences, which stimulated a considerable interest from researchers since the first reports of their fold into non-classical structural motifs known as G-quadruplexes (G4s) [1][2][ (Figure 1A). These structures are characterized by the presence of two or more stacks of four guanines organized in a coplanar manner [4]. Each set of four guanines forms a building block, usually called G-tetrad that are stabilized by Hoogsteen hydrogen base-pairing in physiological conditions, π-π interactions and as well as the presence of positively charged monovalent cations (usually K+ and Na+) (Figure 1B) [5]. G4s are highly polymorphic and can adopt a wide variety of structures based on strand molecularity, strand direction, as well as length and loop composition [6]. According to molecularity, the structures may be distinguished in intramolecular or intermolecular[6]. Considering the direction of the strands, G4 structures may be classified as parallel, anti-parallel and hybrid (Figure 1C-H). The loops are generally divided into three main groups: propeller, lateral, and diagonal [6]. Recently, some structural studies demonstrated the formation of G4 structures with longer loop lengths and bulges, opening the framework for the development of novel diagnostic and therapeutic approaches based on those features[7][8].
Figure 1. (A) Guanine-rich sequence with potential to form a three-tetrad G4. (B) Chemical structure of G-tetrad formed by the Hoogsteen hydrogen-bonded guanines and central cation (colored in gray) coordinated to oxygen atoms. Schematic representation of common unimolecular G4s based on the strand direction: (C) parallel, (D) anti-parallel, and (E) hybrid. Representative PDB structures of (F) parallel (PDB ID: 2M4P), (G) anti-parallel (PDB ID: 1I34) and (H) hybrid (2JPZ) G4 structures. The different loops (propeller, diagonal and lateral) and a bulge were also showed.
Computational algorithms were developed to predict the location of specific G4 sequence motifs in the human genome[9][10]. Such predictors consist on the general motif G≥3NxG≥3NxG≥3NxG≥3 and identified over 370.000 sequences with the potential to fold into G4 structures [11]. However, early algorithms are not accurate and lack the flexibility to accommodate divergences from the canonical pattern. In order to surpass these disadvantages, novel approaches were developed to compute the G4 propensity score by quantifying G-richness (reflecting the fraction of guanines in the sequence) and G-skewness (reflecting G/C asymmetry between the complementary nucleic acid strands) of a given sequence[12][13], or by summing the binding affinities of smaller regions within the G4 and penalizing with the destabilizing effect of loops [14]. Recently, new machine learning approaches were employed to map active G4s based on sequence features and trained using newly available genome-wide mapping of G4s in vitro and in vivo [15][16].
2. G4-seq in high-throughput sequencing methods
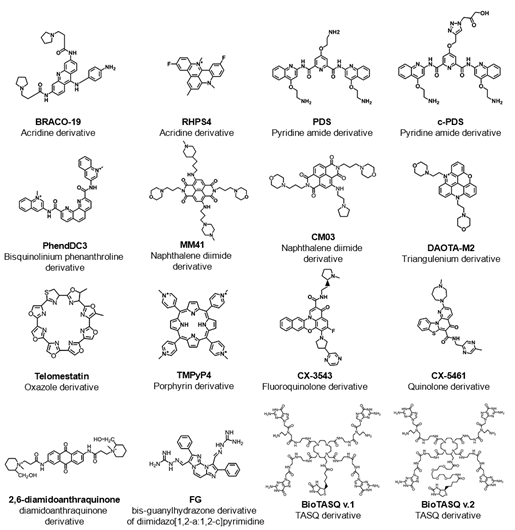
In the last years, the development of high-throughput sequencing methods, such as G4-seq, enables the identification of over 716.000 DNA guanine-rich sequences, across the human genome, with the ability to fold into G4 structures in the presence of the well-known G4 ligand, pyridostatin (PDS) (Figure 2) [17]. PDS has an important role in next-generation sequencing (NGS), since stabilizes G4s and induces polymerase stalling. Those DNA guanine-rich sequences are non-randomly distributed and are mainly located in clusters of immunoglobulin switch regions [18], telomeres [19] and promoter regions of oncogenes [20]. Several reports have described the formation of G4 structures within endogenous chromatin, and their ability to recruit transcription factors to promote active transcription[21[22][23][24][25][26][27][28]. The location of those G4 structures was revealed using an antibody-based G4 chromatin immunoprecipitation sequencing (G4 ChIP–seq) approach [21], and suggest that they play a crucial role in critical cellular processes such as DNA replication [29][30], DNA damage repair [26], transcription [22][, translation[31] and epigenetic modifications [32]. By using G4 ChIP–seq, Hänsel-Hertsch et. al showed a reduction in the number of detected DNA G4s (10.000) in genome [21]. These results are not surprising since transient G4 structures strongly depend on chromatin relaxation and cell status [21]. Recently, an improved version of the G4-seq method was developed and makes available the G4 map of 12 different species [33].
RNA guanine-rich sequences came into the trend of research in the last few years due to their intrinsic features and strengths. RNA G4s are more compact, less hydrated, and more thermodynamically stable than their DNA counterparts [34]. Furthermore, the presence of the 2′-OH group in the ribose ring favors the parallel topology, making them more attractive as target molecules [34]. To date, using computational approaches, more than 1.1 million guanine-rich sequences were identified with the ability to fold into RNA G4[34] . RNA G4s were shown to exist in human cells by using the specific G4 antibody BG4 [36]and like DNA G4s, those sequences are non-randomly distributed in transcriptome [37]. Those sequences are mainly located in both 5′ and 3′UTR, as well as splicing junction of mRNA and noncoding RNAs, being of utmost importance in regulatory post-transcriptional mechanisms [37]. In the last few years, several reports have highlighted the importance of G4s in the transcriptome by employing G4 sequencing high-throughput approaches [38][39][40][41]. rG4-seq was initially applied to map G4s in RNA extracted from HeLa cells [38] and later to plants [40] and bacteria [41]. G4RP-seq was also used to in vivo characterize the G4 transcriptomic landscape [39]. Yang et. al developed a biotinylated template-assembled synthetic G-quartet (TASQ) derivative (BioTASQ v.1) (Figure 2) and captured G4 RNAs from breast cancer cells in log-phase growth, followed by target identification by sequencing [39]. The effect of BRACO-19 and RHPS4 (Figure 2) treatment was also evaluated[39] . They found that those ligands can change the G4 transcriptome in a more remarkable way in long non-coding RNAs [39]. More recently, the same research group developed a new BioTASQ prototype that they called BioTASQ v.2 (Figure 2) and performed an in-depth study of both ligands [42]. Those studies are of utmost importance and revealed the strong relevance that G4 ligands could have in cell biology.
Therefore, the location of G4s at both DNA and RNA levels suggests an active role in the development of diseases like cancer and neurological disorders [43]. Several pieces of evidence suggest that G4s play an important role in promoting genomic instability by triggering DNA damage[44][45][46]. The G4 ligand PDS induces DNA damage as shown by the formation of γH2AX foci, a marker of double-stranded DNA breakage (DSB) [47]. Furthermore, ChIP-seq has shown that PDS accumulates at genes containing clusters of G4 structures and that accumulation is transcription-dependent [44]. Recently, De Magis et. al showed that G4 ligands PDS, BRACO-19 and bis-guanylhydrazone derivative of diimidazo[1,2-a:1,2-c]pyrimidine 1 (FG) (Figure 2) induced the formation of R-loops, another noncanonical secondary of a DNA:RNA hybrid compatible with the formation of a G4, and promote DNA damage as a consequence of that formation [47]. They also found that the mechanism of genome instability and cell killing by G4 ligands was particularly efficient in BRCA2-depleted cancer cells [44]. This study could open up new possibilities of investigation and lead to the development of new anticancer approaches.
Although G4s present in eukaryotic species has been extensively studied, their presence in bacteria and viruses only have gained attention in the last few years [48][49][50][51][52]. In bacteria, G4s are found in regulatory regions that play important functions in replication, radioresistance, antigenic variation and latency [51]. G4s in virus have important regulatory roles in key viral steps [53]. Recent studies have demonstrated the formation and function of G4s in pathogens responsible for serious diseases. Among them are Mycobacterium tuberculosis [54], Pseudomonas aeruginosa [41], Human Papilloma Virus (HPV) [55], Human Immunodeficiency Virus (HIV) [53] and SARS-CoV-2 [56].
Therefore, the recognition of the biological significance of G4s has promoted the research and development of ligands that interact with G4s and regulate their structure and function. The most well-known G4 ligands have been initially developed to target DNA G4s, but many of them have also been employed in the target of RNA G4s [57]. Despite some significant progress in the field, the main challenge remains on the trade-off between affinity and selectivity, which could be achieved with the full characterization of G4/ligand interactions. Since the discovery of the first G4 ligands (disubstituted amidoanthraquinones) (Figure 2) [58], methods like circular dichroism, surface plasmon resonance, isothermal titration calorimetry, mass spectrometry, nuclear magnetic resonance and X-ray crystallography have been used to characterize the molecular interactions of the G4/ligand pair. However, despite the utility of those methods, they are in general, time-consuming and/or costly for the first screening of G4/ligand interactions. Following the general tendency, high-throughput approaches like FRET-melting, G4-Fluorescence intercalator displacement (G4-FID), affinity chromatography and microarrays have emerged as rapid and efficient methods to detect the binding and interaction of ligands with their G4 targets.
Overall, this review describes the most known G4 ligands and highlights the importance of the last developed experimental methods to characterize G4/ligand complex interactions.
Figure 2. List of some examples of G4-interacting ligands mentioned in this review showing the common name of the ligand, chemical structure and family of the compound (chemical backbone).
References
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [CrossRef]
- Arnott, S.; Chandrasekaran, K.; Marttila, C.M. Structures for polyinosinic acid and polyguanylic acid. Biochem. J. 1974, 141, 537–543. [Google Scholar] [CrossRef]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef]
- Largy, E.; Mergny, J.L.; Gabelica, V. Role of Alkali Metal Ions in G-Quadruplex Nucleic Acid Structure and Stability. In Metal Ions in Life Sciences; Springer: Berlin, Germany, 2016; Volume 16, pp. 203–258. [Google Scholar]
- Ma, Y.; Iida, K.; Nagasawa, K. Topologies of G-quadruplex: Biological functions and regulation by ligands. Biochem. Biophys. Res. Commun. 2020, 531, 3–17. [Google Scholar] [CrossRef]
- Ngoc Nguyen, T.Q.; Lim, K.W.; Phan, A.T. Duplex formation in a G-quadruplex bulge. Nucleic Acids Res. 2020, 48, 10567–10575. [Google Scholar] [CrossRef]
- Meier, M.; Moya-Torres, A.; Krahn, N.J.; McDougall, M.D.; Orriss, G.L.; McRae, E.K.S.; Booy, E.P.; McEleney, K.; Patel, T.R.; McKenna, S.A.; et al. Structure and hydrodynamics of a DNA G-quadruplex with a cytosine bulge. Nucleic Acids Res. 2018, 46, 5319–5331. [Google Scholar] [CrossRef]
- Huppert, J.L.; Balasubramanian, S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. [Google Scholar] [CrossRef] [PubMed]
- Huppert, J.L.; Balasubramanian, S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007, 35, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Maizels, N.; Gray, L.T. The G4 Genome. PLoS Genet. 2013, 9, e1003468. [Google Scholar] [CrossRef] [PubMed]
- Bedrat, A.; Lacroix, L.; Mergny, J.-L. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016, 44, 1746–1759. [Google Scholar] [CrossRef]
- Brázda, V.; Kolomazník, J.; Lýsek, J.; Bartas, M.; Fojta, M.; Šťastný, J.; Mergny, J.L. G4Hunter web application: A web server for G-quadruplex prediction. Bioinformatics 2019, 35, 3493–3495. [Google Scholar] [CrossRef] [PubMed]
- Hon, J.; Martínek, T.; Zendulka, J.; Lexa, M. Pqsfinder: An exhaustive and imperfection-tolerant search tool for potential quadruplex-forming sequences in R. Bioinformatics 2017, 33, 3373–3379. [Google Scholar] [CrossRef]
- Klimentova, E.; Polacek, J.; Simecek, P.; Alexiou, P. Penguinn: Precise Exploration of Nuclear G-Quadruplexes Using Interpretable Neural Networks. Front. Genet. 2020, 11, 1287. [Google Scholar] [CrossRef]
- Garant, J.M.; Perreault, J.P.; Scott, M.S. Motif independent identification of potential RNA G-quadruplexes by G4RNA screener. Bioinformatics 2017, 33, 3532–3537. [Google Scholar] [CrossRef]
- Chambers, V.S.; Marsico, G.; Boutell, J.M.; Di Antonio, M.; Smith, G.P.; Balasubramanian, S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015, 33, 877–881. [Google Scholar] [CrossRef]
- Sen, D.; Gilbert, W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 1988, 334, 364–366. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M. G-quadruplexes at telomeres: Friend or foe? Molecules 2020, 25, 3686. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Hänsel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016, 48, 1267–1272. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, J.; Cuesta, S.M.; Adhikari, S.; Hänsel-Hertsch, R.; Tannahill, D.; Balasubramanian, S. G-quadruplexes are transcription factor binding hubs in human chromatin. Genome Biol. 2021, 22, 117. [Google Scholar] [CrossRef] [PubMed]
- Lago, S.; Nadai, M.; Cernilogar, F.M.; Kazerani, M.; Domíniguez Moreno, H.; Schotta, G.; Richter, S.N. Promoter G-quadruplexes and transcription factors cooperate to shape the cell type-specific transcriptome. Nat. Commun. 2021, 12, 3885. [Google Scholar] [CrossRef] [PubMed]
- Jara-Espejo, M.; Peres Line, S.R. DNA G-quadruplex stability, position and chromatin accessibility are associated with CpG island methylation. FEBS J. 2020, 287, 483–495. [Google Scholar] [CrossRef]
- Shen, J.; Varshney, D.; Simeone, A.; Zhang, X.; Adhikari, S.; Tannahill, D.; Balasubramanian, S. Promoter G-quadruplex folding precedes transcription and is controlled by chromatin. Genome Biol. 2021, 22, 143. [Google Scholar] [CrossRef]
- Komůrková, D.; Kovaříková, A.S.; Bártová, E. G-quadruplex structures colocalize with transcription factories and nuclear speckles surrounded by acetylated and dimethylated histones H3. Int. J. Mol. Sci. 2021, 22, 1995. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, R.F.; Moshkin, Y.M.; Mouton, S.; Grzeschik, N.A.; Kalicharan, R.D.; Kuipers, J.; Wolters, A.H.G.; Nishida, K.; Romashchenko, A.V.; Postberg, J.; et al. Guanine quadruplex structures localize to heterochromatin. Nucleic Acids Res. 2016, 44, 152–163. [Google Scholar] [CrossRef]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef]
- Lee, W.T.C.; Yin, Y.; Morten, M.J.; Tonzi, P.; Gwo, P.P.; Odermatt, D.C.; Modesti, M.; Cantor, S.B.; Gari, K.; Huang, T.T.; et al. Single-molecule imaging reveals replication fork coupled formation of G-quadruplex structures hinders local replication stress signaling. Nat. Commun. 2021, 12, 2525. [Google Scholar] [CrossRef]
- Tran, P.L.T.; Rieu, M.; Hodeib, S.; Joubert, A.; Ouellet, J.; Alberti, P.; Bugaut, A.; Allemand, J.F.; Boulé, J.B.; Croquette, V. Folding and persistence times of intramolecular G-quadruplexes transiently embedded in a DNA duplex. Nucleic Acids Res. 2021, 49, 5189–5201. [Google Scholar] [CrossRef]
- Murat, P.; Marsico, G.; Herdy, B.; Ghanbarian, A.; Portella, G.; Balasubramanian, S. RNA G-quadruplexes at upstream open reading frames cause DHX36- and DHX9-dependent translation of human mRNAs. Genome Biol. 2018, 19, 229. [Google Scholar] [CrossRef]
- Dutta, A.; Maji, N.; Sengupta, P.; Banerjee, N.; Kar, S.; Mukherjee, G.; Chatterjee, S.; Basu, M. Promoter G-quadruplex favours epigenetic reprogramming-induced atypical expression of ZEB1 in cancer cells. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129899. [Google Scholar] [CrossRef] [PubMed]
- Marsico, G.; Chambers, V.S.; Sahakyan, A.B.; McCauley, P.; Boutell, J.M.; Antonio, M.D.; Balasubramanian, S. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 2019, 47, 3862–3874. [Google Scholar] [CrossRef]
- Zaccaria, F.; Fonseca Guerra, C. RNA versus DNA G-Quadruplex: The Origin of Increased Stability. Chem. Eur. J. 2018, 24, 16315–16322. [Google Scholar] [CrossRef] [PubMed]
- Vannutelli, A.; Belhamiti, S.; Garant, J.-M.; Ouangraoua, A.; Perreault, J.-P. Where are G-quadruplexes located in the human transcriptome? NAR Genomics Bioinform. 2020, 2, lqaa035. [Google Scholar] [CrossRef]
- Biffi, G.; Di Antonio, M.; Tannahill, D.; Balasubramanian, S. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 2014, 6, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Tassinari, M.; Richter, S.N.; Gandellini, P. Biological relevance and therapeutic potential of G-quadruplex structures in the human noncoding transcriptome. Nucleic Acids Res. 2021, 49, 3617–3633. [Google Scholar] [CrossRef]
- Kwok, C.K.; Marsico, G.; Sahakyan, A.B.; Chambers, V.S.; Balasubramanian, S. RG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Methods 2016, 13, 841–844. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Y.; Lejault, P.; Chevrier, S.; Boidot, R.; Robertson, A.G.; Wong, J.M.Y.; Monchaud, D. Transcriptome-wide identification of transient RNA G-quadruplexes in human cells. Nat. Commun. 2018, 9, 4730. [Google Scholar] [CrossRef]
- Yang, X.; Cheema, J.; Zhang, Y.; Deng, H.; Duncan, S.; Umar, M.I.; Zhao, J.; Liu, Q.; Cao, X.; Kwok, C.K.; et al. RNA G-quadruplex structures exist and function in vivo in plants. Genome Biol. 2020, 21, 226. [Google Scholar] [CrossRef]
- Shao, X.; Zhang, W.; Umar, M.I.; Wong, H.Y.; Seng, Z.; Xie, Y.; Zhang, Y.; Yang, L.; Kwok, C.K.; Deng, X. RNA G-quadruplex structures mediate gene regulation in bacteria. MBio 2020, 11, e02926-19. [Google Scholar] [CrossRef]
- Renard, I.; Grandmougin, M.; Roux, A.; Yang, S.Y.; Lejault, P.; Pirrotta, M.; Wong, J.M.Y.; Monchaud, D. Small-molecule affinity capture of DNA/RNA quadruplexes and their identification in vitro and in vivo through the G4RP protocol. Nucleic Acids Res. 2019, 47, 502–510. [Google Scholar] [CrossRef]
- Maizels, N. G4-associated human diseases. EMBO Rep. 2015, 16, 910–922. [Google Scholar] [CrossRef] [PubMed]
- De Magis, A.; Manzo, S.G.; Russo, M.; Marinello, J.; Morigi, R.; Sordet, O.; Capranico, G. DNA damage and genome instability by G-quadruplex ligands are mediated by R loops in human cancer cells. Proc. Natl. Acad. Sci. USA 2019, 116, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Zell, J.; Sperti, F.R.; Britton, S.; Monchaud, D. DNA folds threaten genetic stability and can be leveraged for chemotherapy. RSC Chem. Biol. 2021, 2, 47–76. [Google Scholar] [CrossRef]
- Maffia, A.; Ranise, C.; Sabbioneda, S. From R-loops to G-quadruplexes: Emerging new threats for the replication fork. Int. J. Mol. Sci. 2020, 21, 1506. [Google Scholar] [CrossRef]
- Rodriguez, R.; Miller, K.M.; Forment, J.V.; Bradshaw, C.R.; Nikan, M.; Britton, S.; Oelschlaegel, T.; Xhemalce, B.; Balasubramanian, S.; Jackson, S.P. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012, 8, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Yadav, P.; Kim, N.; Kumari, M.; Verma, S.; Sharma, T.K.; Yadav, V.; Kumar, A. G-quadruplex structures in bacteria: Biological relevance and potential as an antimicrobial target. J. Bacteriol. 2021, 203, e0057720. [Google Scholar] [CrossRef]
- Ruggiero, E.; Richter, S.N. Viral G-quadruplexes: New frontiers in virus pathogenesis and antiviral therapy. In Annual Reports in Medicinal Chemistry; Elsevier: Amsterdam, The Netherlands, 2020; Volume 54, pp. 101–131. [Google Scholar]
- Abiri, A.; Lavigne, M.; Rezaei, M.; Nikzad, S.; Zare, P.; Mergny, J.L.; Rahimi, H.R. Unlocking G-quadruplexes as antiviral targets. Pharmacol. Rev. 2021, 73, 897–923. [Google Scholar] [CrossRef]
- Saranathan, N.; Vivekanandan, P. G-Quadruplexes: More Than Just a Kink in Microbial Genomes. Trends Microbiol. 2019, 27, 148–163. [Google Scholar] [CrossRef]
- Seifert, H.S. Above and beyond Watson and Crick: Guanine Quadruplex Structures and Microbes. Annu. Rev. Microbiol. 2018, 72, 49–69. [Google Scholar] [CrossRef]
- Metifiot, M.; Amrane, S.; Litvak, S.; Andreola, M.L. G-quadruplexes in viruses: Function and potential therapeutic applications. Nucleic Acids Res. 2014, 42, 12352–12366. [Google Scholar] [CrossRef]
- Perrone, R.; Lavezzo, E.; Riello, E.; Manganelli, R.; Palù, G.; Toppo, S.; Provvedi, R.; Richter, S.N. Mapping and characterization of G-quadruplexes in Mycobacterium tuberculosis gene promoter regions. Sci. Rep. 2017, 7, 5743. [Google Scholar] [CrossRef]
- Tlučková, K.; Marušič, M.; Tóthová, P.; Bauer, L.; Šket, P.; Plavec, J.; Viglasky, V. Human papillomavirus G-quadruplexes. Biochemistry 2013, 52, 7207–7216. [Google Scholar] [CrossRef]
- Zhao, C.; Qin, G.; Niu, J.; Wang, Z.; Wang, C.; Ren, J.; Qu, X. Targeting RNA G-Quadruplex in SARS-CoV-2: A Promising Therapeutic Target for COVID-19? Angew. Chem. Int. Ed. 2021, 60, 432–438. [Google Scholar] [CrossRef]
- Asamitsu, S.; Obata, S.; Yu, Z.; Bando, T.; Sugiyama, H. Recent progress of targeted G-quadruplex-preferred ligands toward cancer therapy. Molecules 2019, 24, 429. [Google Scholar] [CrossRef]
- Sun, D.; Thompson, B.; Cathers, B.E.; Salazar, M.; Kerwin, S.M.; Trent, J.O.; Jenkins, T.C.; Neidle, S.; Hurley, L.H. Inhibition of human telomerase by a G-Quadruplex-Interactive compound. J. Med. Chem. 1997, 40, 2113–2116. [Google Scholar] [CrossRef]