Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Dimitrios S. Kanakoglou | + 2051 word(s) | 2051 | 2021-11-15 08:03:12 | | | |
| 2 | Nora Tang | Meta information modification | 2051 | 2021-12-01 10:22:18 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Kanakoglou, D. Main Characteristics of CircRNAs. Encyclopedia. Available online: https://encyclopedia.pub/entry/16563 (accessed on 08 February 2026).
Kanakoglou D. Main Characteristics of CircRNAs. Encyclopedia. Available at: https://encyclopedia.pub/entry/16563. Accessed February 08, 2026.
Kanakoglou, Dimitrios. "Main Characteristics of CircRNAs" Encyclopedia, https://encyclopedia.pub/entry/16563 (accessed February 08, 2026).
Kanakoglou, D. (2021, November 30). Main Characteristics of CircRNAs. In Encyclopedia. https://encyclopedia.pub/entry/16563
Kanakoglou, Dimitrios. "Main Characteristics of CircRNAs." Encyclopedia. Web. 30 November, 2021.
Copy Citation
Circular RNAs (circRNAs) represent a category of non-coding RNAs (ncRNAs) and constitute functional RNAs that are predetermined not to be translated, but transcribed. CircRNAs are characterized by single-stranded closed-loop structures without 5′-3′-polarity or a polyadenylated tail. The term “circRNA” was first used by Sanger et al. when identifying the structure of viroids.
circular RNAs
CircRNAs
1. Categories, Biogenesis, Localization, Degradation
CircRNAs are categorized according to their synthesis by introns, exons, or both into four main categories described below:
-
Intronic circRNAs (ciRNAs), localized to the cell nucleus [4].
-
Exon-Intron circRNAs (EIcircRNAs), also localized to the cell nucleus and functionally similar to ciRNAs [5].
-
Intergenic circRNAs, generated by two ciRNAs fragments flanked by GT-AC splicing signals, acting as the splice donor and acceptor of the circular junction, and forming an integrated circRNAs [6].
CircRNAs are generated by back-splicing (Figure 1) of primary messenger RNA (mRNA) transcripts. During back-splicing, a 5′ splice site (donor) is joined to an upstream 3′ site (acceptor) by the spliceosome machinery, forming a covalently closed structure with a specific junction site [4]. This process competes with canonical splicing and can produce different circRNAs from the same sequence [7][8]. It has been reported that the more back-splicing an exon can undergo, the less it is included in the fully processed mRNA [9].
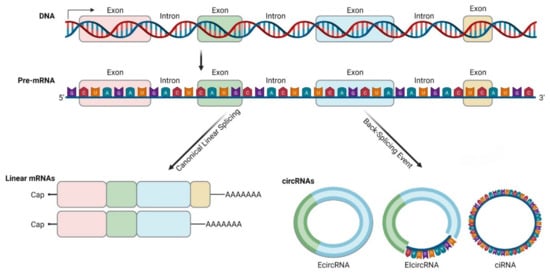
Figure 1. Biogenesis and diversity of circRNAs. CircRNAs are produced by exon-skipping or another non-canonical event (like back-splicing) that can initiate internal splicing. CircRNAs are present in blood, body fluids and tissues, and can serve as potential disease biomarkers. They are non-polyadenylated, unlike mRNAs, and they are covalently closed. CircRNAs are resistant to exonucleases digestion due to their lack of 5′ or 3′ end. CircRNAs can be exonic, intronic or a combination of both (this illustration was created with BioRender.com, accessed on 3 November 2021).
Back-splicing occurs both co-transcriptionally and post-transcriptionally and is a much less efficient process than canonical splicing. It is favored over canonical splicing up on depletion of splicing factors and can be affected by epigenetic changes within histones and gene bodies [7][10][11][12].
There are three basic models of circRNAs biogenesis. Firstly, on an exon-skipping event, lariats are formed when alternative exons are spliced out of the final mRNA product. The lariat containing the excised exons undergoes internal back-splicing and circRNA (EcircRNA or EIcircRNA) are formed [9][13]. Intron lariats that contain a ciRNA specific consensus motif consisting of an 11-Nucleotide (nt) C-rich element near the branch point and a 7-nt GU-rich element near the 5′ splice site can escape debranching and form ciRNAs according to this model [14]. Secondly, on “intron pairing-driven circularization”, inverted repeat elements (for example ALU elements) are located in the upstream and downstream introns. By base-pairing of complementary sequences of inverted repeat elements, a hairpin structure is formed and looping brings the acceptor and donor sites in proximity [3][4][9]. Thirdly, looping can be mediated by RNA-Binding Proteins (RBPs), such as Quaking, Muscleblind or FUS/TLS (Fused in Sarcoma/Translocated in Liposarcoma) protein [15][16][17]. Interestingly, not all RBPs promote circRNAs biogenesis, as ADAR1 (Adenosine Deaminase RNA-Specific Binding Protein) binds to double-stranded RNA regions and promotes the melting of stem structures to destabilize RNA pairing, thereby suppressing circRNAs formation [18].
Apart from the four main categories, there are newly emerging subtypes of circRNAs, such as tRNA Intronic circRNAs (tricRNAs), generated during pre-tRNA maturation or fusion circRNAs (f-circRNAs), created from chromosomal translocations in host genes [19][20].
Following synthesis, all circRNAs, apart from intron-containing, are transported from the nucleus to the cytoplasm in a size-dependent manner by the enzymes Adenosine Triphosphate (ATP)-dependent RNA helicase DDX39A and spliceosome RNA helicase DDX39B [21].
First reports regarding the mechanisms implicated on circRNAs degradation were based on studies of the circRNAs Cerebellar Degeneration-Related Protein 1 Transcript (CDR1as), a circRNA upregulated in OS, that described cleavage of CDR1as as mediated by Argonaute 2 (Ago2), after binding to microRNA (miRNA) miR-671 [22]. Nevertheless, whether this type of degradation is universal to circRNAs is still not clear [7]. Recent evidence points to other endonucleases functioning on circRNAs decay, upon special conditions. A portion of circRNAs that have undergone m6A (N6-Methyladenosine) modification are cleaved by the ribonuclease complex RNase P/MRP, a process mediated by the m6A reader protein YTHDF2 (YTH N6-Methyladenosine RNA Binding Protein 2) and HRSP12 (Heat-Responsive Protein 12) [23]. That could be the case for circNRIP1, a circRNA upregulated in OS. Its expression was reportedly elevated by METTL3, a methyltransferase that induces m6A modification to circNRIP1. In turn, circNRIP1 sponges miR-199a to upregulate FOXC2 expression in OS [24]. Additionally, RNase L, activated upon viral infection, can degrade all types of circRNAs. UPF1 (ATP-Dependent RNA Helicase Upstream Frameshift 1) and associated endonuclease G3BP1 (Ras Gtpase-Activating Protein-Binding Protein 1) can target and catalyze the degradation of some circRNAs [25]. Moreover, circRNAs accumulation is eliminated through exocytosis or exosome activity [26].
2. Biochemical Properties and Detection Methods
CircRNAs possess an exceptionally stable structure that results in their accumulation to the cytoplasm as previously mentioned, along with possible mechanisms of degradation. Most EcircRNAs’ half-life surpasses 48 h, in contrast to linear mRNA with average half-life of 10 h [1].
Another feature of circRNAs is their tissue -and developmental stage- or age-specificity. In human tissues, RNA-sequencing studies demonstrated that up to 50% of circRNAs were expressed in a highly specific pattern and showed increased levels on fetal compared to adult tissues [27].
CircRNAs are abundant in human tissues and tend to accumulate in tissues with low proliferation rate, for example cardiomyocytes. CircRNAs accumulation is also dependent on age [28][29]. Different human tissues show variable rates of circRNAs production, as in the human heart where 9% of the expressed genes produce circRNAs, whereas in the human brain the rate of production reaches 20% [30]. However, circRNA expression is not related to that of its linear isoforms, and under specific circumstances can far exceed it [7].
CircRNAs’ stability to degradation and the lack of polar structure at the end are properties that can be utilized for their detection. Their migration rate in a cross-linked gel is slower than that of long linear RNAs, and when compared with homologous gene transcription, nucleic acids show slower migration rate in weakly cross-linked gels, enabling circRNAs detection with northern blot analysis [31][32]. Subcellular location can be assessed with the Fluorescence In Situ Hybridization (FISH) technique [5][14]. Improved algorithms for high-throughput sequencing (circRNA candidate sequence boundary combination concerning different forms of exon rearrangement in comparison to sequencing data, different sequence alignment algorithms to match sequencing data to the genomic sequence and direct detection from cDNA sequences by designing multiple splice sequences) have also rendered the detection of low-abundance circRNAs feasible [33][34]. Currently, multiple circRNA-associated in silico tools and pipelines can support the de novo identification, assembly and annotation of circRNAs [35]. Depending on their implementation, circRNA identification tools are divided into three categories; BSJ-based (Back-Splicing Junction), integrated-based and machine learning-based [36]. Additionally, novel tools can perform more complex functions (e.g., alternative splicing events, expression estimation and circRNA structure prediction). In their review, Chen et al. [36] provide a comprehensive guide for hundreds of circRNA-related tools and their functionality.
3. Biological Functions
CircRNAs exert many important biological functions through three major mechanisms of action (Figure 2):
-
MiRNA sponging
-
Protein and mRNA interplay
-
Translation
MiRNA sponging is the most well-studied mechanism of action of circRNAs. CircRNAs contain binding sites for miRNAs and compete endogenous RNAs to sequester miRNAs via complementary base-pairing, while inhibiting miRNA from binding to their target molecules [37]. Subsequently, miRNA sponging leads to suppression of target mRNAs. The well-known paradigm of circRNA CDR1as, with more than 70 binding sites for miR-7 [38], but also other circRNAs such as circRNA-Homeodomain-Interacting Protein Kinase-2 (circHIPK2), have mechanistically confirmed sponge effect to miR124-2HG [39]. Considering their multiple binding sites, circRNAs can act both as oncogenes or tumor suppressors through binding to different miRNAs [4]. A large part of research focusing on circRNA role on sarcomas refers to circRNA-miRNA interactions and their impact on tumor growth, described later. Nevertheless, bioinformatics analysis reports that although circRNAs may have abundant binding sites, they may not exert prominent sponge effect, and under normal conditions this effect is not important on highly expressed miRNAs [2][22][40][41].
CircRNAs interactions with proteins are multifold, since they can act as protein sponges, scaffolds, enhance protein function or recruit proteins to specific subcellular compartments, or even facilitate contact between two or more proteins. It is well documented that circRNAs that carry RBP binding motifs may act as decoys or sponges to RBPs and interfere with their effects [4]. CircMbl is documented to bind both mbl and MBNL1 (Muscleblind-like Protein 1) proteins. Upon increased mbl or MBNL1 proteins, circMbl synthesis is promoted. In turn, linear mRNA splicing of the gene is favored, creating an autoregulatory loop of the gene expression [15]. In addition, RNA Polymerase II-U1 (Pol II-U1) Small Nuclear Ribonucleoproteins (snRNPs) complex binding is facilitated by interaction with circEIF3J with U1 RNA, resulting in upregulated parental gene transcription [5]. Similarly, circRNAs can bind dsRBPs [Double-Strand RNA-Binding Proteins (e.g., NF90, NF110)] [42] and create a reservoir to be used in specific conditions. Moreover, several circRNAs are reported to act as mRNA decoys or modulate mRNA stability [22][43]. Reports of proteins binding circRNAs without predicted binding sites imply that the tertiary structure of circRNAs may play an important role on protein binding, and that the tertiary structure may exhibit certain fluidity in different developmental stages or tissues [44]. The mechanisms underlying the way that tertiary structure affects circRNA function should be an object of further research.
Bioinformatics have proven that circRNAs have Open Reading Frame (ORF) and ribosome binding site, making translation possible. Translation may occur via Internal Ribosome Entry Site (IRES) elements that recruit the ribosomal 40S subunits in a cap-independent manner [45]. m6A modification may also play a role in circRNA translation, as m6A reader protein YTHDF3 interacts with translation initiation factors elF4G2 and elF3A, after binding to m6A modified circRNA [27]. The resulting peptides are truncated versions of the original proteins and, therefore, their functional relevance remains to be investigated. They might serve as dominant-negative protein variants, having been expressed under different conditions than the original protein or localized to other cellular compartments [45].
It has been mentioned above that circRNAs that localize in the nucleus (EIcircRNAs and ciRNAs) can regulate their parental gene expression by interacting with U1 snRNP complex. CircRNAs also regulate alternative splicing and transcription, given that under certain circumstances (depletion of splicing factors) circRNA biosynthesis is promoted over canonical splicing [15], exerting an important role on translation regulation by interfering with mRNA mobility and stability [22][43][46]. Other studies also show that circRNAs control the role of ribosomes in protein expression [47]. In addition, recent data demonstrate that stabilized circRNAs could form circRNA pseudogenes and be retrotranscribed and integrated into the genome [48][49], thus modifying gene expression.
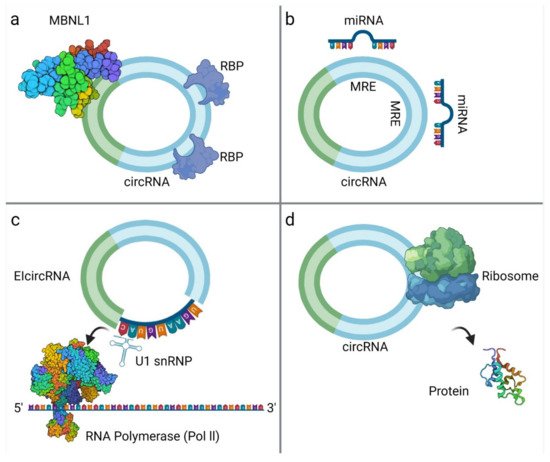
Figure 2. CircRNA interactions: (a) CircRNA binds with RNA-Binding Proteins (RBPs), such as MBNL1 [50]; (b) circRNAs act as miRNA sponges; (c) EIcircRNA interacts with the U1 Small Nuclear Ribonucleoprotein (snRNP) and then binds to RNA Polymerase II (Pol II) [51]; (d) circRNA translation into proteins. The surface of the molecules MBNL1 and Pol II are modelled according to van der Waals surface (this illustration was created with BioRender.com, accessed on 3 November 2021).
4. Roles in Homeostasis and Malignancy
CircRNAs participate in cell cycle regulation as key factors in multiple check points [52] and modulate c-MYC expression as well as other molecules downstream of c-MYC, including p21. They have also been involved in mediation of immune response [53], cell differentiation and pluripotency [54], as well as apoptosis [55].
Recent literature points to the dysregulation of circRNAs in cancer, inducing oncogenic or suppressive effects and playing a pivotal role in tumorigenesis [52]. Bioinformatics analysis has yielded interesting results on many tumor types, identifying a network of circRNAs and their target miRNAs and mRNAs as well as their interaction with important signaling pathways, such as Wnt/β-catenin, TGF-β (Transforming Growth Factor Beta) or PI3K/AKT [52]. They have also been shown to regulate Epithelial-Mesenchymal Transition (EMT), resist apoptosis, induce angiogenesis, DNA methylation and demethylation coordination as well as influence metastatic spread [37][52], whereas some studies have indicated the presence of circRNAs in exosomes, implicating their transport between cells and tissues [7][56][57]. Moreover, new evidence implies that circRNAs may affect acquired chemoresistance [58]. The above-mentioned mechanisms have been shown to be critical in sarcomagenesis [59][60][61][62][63][64][65], and a large number of circRNAs have been demonstrated to affect chemoresistance in OS, such as hsa_circ_0004674, hsa_circ_0081001 and circPVT1, among others [59][66][67][68][69].
References
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157.
- Guo, J.U.; Agarwal, V.; Guo, H.; Bartel, D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014, 15, 409.
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary Sequence-Mediated Exon Circularization. Cell 2014, 159, 134–147.
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691.
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264.
- Gao, Y.; Wang, J.; Zhao, F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015, 16, 1–16.
- Chen, L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016, 17, 205–211.
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733.
- Kelly, S.; Greenman, C.; Cook, P.R.; Papantonis, A. Exon skipping is correlated with exon circularization. J. Mol. Biol. 2015, 427, 2414–2417.
- Shukla, S.; Kavak, E.; Gregory, M.; Imashimizu, M.; Shutinoski, B.; Kashlev, M.; Oberdoerffer, P.; Sandberg, R.; Oberdoerffer, S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 2011, 479, 74–79.
- Bentley, D.L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 2014, 15, 163–175.
- Kramer, M.C.; Liang, D.; Tatomer, D.C.; Gold, B.; March, Z.M.; Cherry, S.; Wilusz, J.E. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015, 29, 2168–2182.
- Eger, N.; Schoppe, L.; Schuster, S.; Laufs, U.; Boeckel, J.N. Circular RNA splicing. Circ. RNAs 2018, 1087, 41–52.
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806.
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66.
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134.
- Errichelli, L.; Modigliani, S.D.; Laneve, P.; Colantoni, A.; Legnini, I.; Capauto, D.; Rosa, A.; De Santis, R.; Scarfo, R.; Peruzzi, G. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun. 2017, 8, 14741.
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.T.; Orejuela, M.R.; Piechotta, M.; Levanon, E.Y.; Landthaler, M.; Dieterich, C. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015, 10, 170–177.
- Alderton, G.K. Circular RNAs from translocations. Nat. Rev. Cancer 2016, 16, 273.
- Guarnerio, J.; Bezzi, M.; Jeong, J.C.; Paffenholz, S.V.; Berry, K.; Naldini, M.M.; Lo-Coco, F.; Tay, Y.; Beck, A.H.; Pandolfi, P.P. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell 2016, 165, 289–302.
- Huang, C.; Liang, D.; Tatomer, D.C.; Wilusz, J.E. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018, 32, 639–644.
- Hansen, T.B.; Wiklund, E.D.; Bramsen, J.B.; Villadsen, S.B.; Statham, A.L.; Clark, S.J.; Kjems, J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011, 30, 4414–4422.
- Park, O.H.; Ha, H.; Lee, Y.; Boo, S.H.; Kwon, D.H.; Song, H.K.; Kim, Y.K. Endoribonucleolytic cleavage of m6A-containing RNAs by RNase P/MRP complex. Mol. Cell 2019, 74, 494–507.e8.
- Meng, Y.; Hao, D.; Huang, Y.; Jia, S.; Zhang, J.; He, X.; Liu, D.; Sun, L. Circular RNA circNRIP1 plays oncogenic roles in the progression of osteosarcoma. Mamm. Genome 2021.
- Fischer, J.W.; Busa, V.F.; Shao, Y.; Leung, A.K. Structure-mediated RNA decay by UPF1 and G3BP1. Mol. Cell 2020, 78, 70–84.
- Preußer, C.; Hung, L.H.; Schneider, T.; Schreiner, S.; Hardt, M.; Moebus, A.; Santoso, S.; Bindereif, A. Selective release of circRNAs in platelet-derived extracellular vesicles. J. Extracell. Vesicles 2018, 7, 1424473.
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017, 27, 626–641.
- Du, W.W.; Yang, W.; Chen, Y.; Wu, Z.K.; Foster, F.S.; Yang, Z.; Li, X.; Yang, B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017, 38, 1402–1412.
- Bachmayr-Heyda, A.; Reiner, A.T.; Auer, K.; Sukhbaatar, N.; Aust, S.; Bachleitner-Hofmann, T.; Mesteri, I.; Grunt, T.W.; Zeillinger, R.; Pils, D. Correlation of circular RNA abundance with proliferation—Exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis and normal human tissues. Sci. Rep. 2015, 5, 8057.
- Aufiero, S.; Reckman, Y.J.; Pinto, Y.M.; Creemers, E.E. Circular RNAs open a new chapter in cardiovascular biology. Nat. Rev. Cardiol. 2019, 16, 503–514.
- Suzuki, H.; Tsukahara, T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int. J. Mol. Sci. 2014, 15, 9331–9342.
- Tabak, H.F.; Van der Horst, G.; Smit, J.; Winter, A.J.; Mul, Y.; Groot Koerkamp, M.J. Discrimination between RNA circles, interlocked RNA circles and lariats using two-dimensional polyacrylamide gel electrophoresis. Nucleic Acids Res. 1988, 16, 6597–6605.
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-Type Specific Features of Circular RNA Expression. PLoS Genet. 2013, 9, e1003777.
- Hoffmann, S.; Otto, C.; Doose, G.; Tanzer, A.; Langenberger, D.; Christ, S.; Kunz, M.; Holdt, L.M.; Teupser, D.; Hackermüller, J.; et al. A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol. 2014, 15, R34.
- Zeng, X.; Lin, W.; Guo, M.; Zou, Q. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput. Biol. 2017, 13, e1005420.
- Chen, L.; Wang, C.; Sun, H.; Wang, J.; Liang, Y.; Wang, Y.; Wong, G. The bioinformatics toolbox for circRNA discovery and analysis. Brief. Bioinform. 2021, 22, 1706–1728.
- Ng, W.L.; Mohd Mohidin, T.B.; Shukla, K. Functional role of circular RNAs in cancer development and progression. RNA Biol. 2018, 15, 995–1005.
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388.
- Huang, R.; Zhang, Y.; Han, B.; Bai, Y.; Zhou, R.; Gan, G.; Chao, J.; Hu, G.; Yao, H. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124–2HG. Autophagy 2017, 13, 1722–1741.
- You, X.; Vlatkovic, I.; Babic, A.; Will, T.; Epstein, I.; Tushev, G.; Akbalik, G.; Wang, M.; Glock, C.; Quedenau, C.; et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015, 18, 603–610.
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283.
- Li, X.; Liu, C.X.; Xue, W.; Zhang, Y.; Jiang, S.; Yin, Q.F.; Wei, J.; Yao, R.W.; Yang, L.; Chen, L.L. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol. Cell 2017, 67, 214–227.e217.
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461.
- Du, W.W.; Zhang, C.; Yang, W.; Yong, T.; Awan, F.M.; Yang, B.B. Identifying and Characterizing circRNA-Protein Interaction. Theranostics 2017, 7, 4183–4191.
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 2017, 66, 22–37.e29.
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379.
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 12429.
- Liu, J.; Liu, T.; Wang, X.; He, A. Circles reshaping the RNA world: From waste to treasure. Mol. Cancer 2017, 16, 58.
- Xu, S.; Zhou, L.; Ponnusamy, M.; Zhang, L.; Dong, Y.; Zhang, Y.; Wang, Q.; Liu, J.; Wang, K. A comprehensive review of circRNA: From purification and identification to disease marker potential. PeerJ 2018, 6, e5503.
- Park, S.; Phukan, P.D.; Zeeb, M.; Martinez-Yamout, M.A.; Dyson, H.J.; Wright, P.E. Structural Basis for Interaction of the Tandem Zinc Finger Domains of Human Muscleblind with Cognate RNA from Human Cardiac Troponin T. Biochemistry 2017, 56, 4154–4168.
- Jishage, M.; Yu, X.; Shi, Y.; Ganesan, S.J.; Chen, W.Y.; Sali, A.; Chait, B.T.; Asturias, F.J.; Roeder, R.G. Architecture of Pol II(G) and molecular mechanism of transcription regulation by Gdown1. Nat. Struct. Mol. Biol. 2018, 25, 859–867.
- Li, J.; Sun, D.; Pu, W.; Wang, J.; Peng, Y. Circular RNAs in Cancer: Biogenesis, Function, and Clinical Significance. Trends Cancer 2020, 6, 319–336.
- Chen, Y.G.; Kim, M.V.; Chen, X.; Batista, P.J.; Aoyama, S.; Wilusz, J.E.; Iwasaki, A.; Chang, H.Y. Sensing Self and Foreign Circular RNAs by Intron Identity. Mol. Cell 2017, 67, 228–238.e225.
- Yu, C.Y.; Li, T.C.; Wu, Y.Y.; Yeh, C.H.; Chiang, W.; Chuang, C.Y.; Kuo, H.C. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat. Commun. 2017, 8, 1149.
- Zhang, H.; Wang, G.; Ding, C.; Liu, P.; Wang, R.; Ding, W.; Tong, D.; Wu, D.; Li, C.; Wei, Q.; et al. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget 2017, 8, 61687–61697.
- Shen, B.; Wang, Z.; Li, Z.; Song, H.; Ding, X. Circular RNAs: An emerging landscape in tumor metastasis. Am. J. Cancer Res. 2019, 9, 630.
- Wang, Y.; Liu, J.; Ma, J.; Sun, T.; Zhou, Q.; Wang, W.; Wang, G.; Wu, P.; Wang, H.; Jiang, L.; et al. Exosomal circRNAs: Biogenesis, effect and application in human diseases. Mol. Cancer 2019, 18, 116.
- Kartal-Yandim, M.; Adan-Gokbulut, A.; Baran, Y. Molecular mechanisms of drug resistance and its reversal in cancer. Crit. Rev. Biotechnol. 2016, 36, 716–726.
- Bai, Y.; Li, Y.; Bai, J.; Zhang, Y. Hsa_circ_0004674 promotes osteosarcoma doxorubicin resistance by regulating the miR-342-3p/FBN1 axis. J. Orthop. Surg. Res. 2021, 16, 510.
- Zhang, H.; Yan, J.; Lang, X.; Zhuang, Y. Expression of circ_001569 is upregulated in osteosarcoma and promotes cell proliferation and cisplatin resistance by activating the Wnt/β-catenin signaling pathway. Oncol. Lett. 2018, 16, 5856–5862.
- Wu, Z.; Shi, W.; Jiang, C. Overexpressing circular RNA hsa_circ_0002052 impairs osteosarcoma progression via inhibiting Wnt/β-catenin pathway by regulating miR-1205/APC2 axis. Biochem. Biophys. Res. Commun. 2018, 502, 465–471.
- Wang, L.; Du, Z.G.; Huang, H.; Li, F.S.; Li, G.S.; Xu, S.N. Circ-0003998 promotes cell proliferative ability and invasiveness by binding to miR-197-3p in osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10638–10646.
- Zhang, Z.; Pu, F.; Wang, B.; Wu, Q.; Liu, J.; Shao, Z. Hsa_circ_0000285 functions as a competitive endogenous RNA to promote osteosarcoma progression by sponging hsa-miRNA-599. Gene Ther. 2020, 27, 186–195.
- Seo, T.; Park, J.; Lee, D.; Hwang Sun, G.; Choe, J. Viral Interferon Regulatory Factor 1 of Kaposi’s Sarcoma-Associated Herpesvirus Binds to p53 and Represses p53-Dependent Transcription and Apoptosis. J. Virol. 2001, 75, 6193–6198.
- Shin Young, C.; Nakamura, H.; Liang, X.; Feng, P.; Chang, H.; Kowalik Timothy, F.; Jung Jae, U. Inhibition of the ATM/p53 Signal Transduction Pathway by Kaposi’s Sarcoma-Associated Herpesvirus Interferon Regulatory Factor 1. J. Virol. 2006, 80, 2257–2266.
- Zhu, K.-P.; Zhang, C.-L.; Hu, J.-P.; Zhang, L. A novel circulating hsa_circ_0081001 act as a potential biomarker for diagnosis and prognosis of osteosarcoma. Int. J. Biol. Sci. 2018, 14, 1513–1520.
- Zhu, K.-P.; Ma, X.-L.; Zhang, L.; Zhang, C.-L.; Hu, J.-P.; Zhan, T.-C. Screening circular RNA related to chemotherapeutic resistance in osteosarcoma by RNA sequencing. Epigenomics 2018, 10, 1327–1346.
- Wei, W.; Ji, L.; Duan, W.; Zhu, J. Circular RNA circ_0081001 knockdown enhances methotrexate sensitivity in osteosarcoma cells by regulating miR-494-3p/TGM2 axis. J. Orthop. Surg. Res. 2021, 16, 50.
- Li, D.; Huang, Y.; Wang, G. Circular RNA circPVT1 Contributes to Doxorubicin (DXR) Resistance of Osteosarcoma Cells by Regulating TRIAP1 via miR-137. Biomed Res. Int. 2021, 2021, 7463867.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.3K
Revisions:
2 times
(View History)
Update Date:
06 Jan 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




