Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Heather Stout-Delgado | + 2796 word(s) | 2796 | 2021-11-19 06:57:42 | | | |
| 2 | Rita Xu | Meta information modification | 2796 | 2021-12-01 02:50:06 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Stout-Delgado, H. Antiviral Gene Expression. Encyclopedia. Available online: https://encyclopedia.pub/entry/16562 (accessed on 07 February 2026).
Stout-Delgado H. Antiviral Gene Expression. Encyclopedia. Available at: https://encyclopedia.pub/entry/16562. Accessed February 07, 2026.
Stout-Delgado, Heather. "Antiviral Gene Expression" Encyclopedia, https://encyclopedia.pub/entry/16562 (accessed February 07, 2026).
Stout-Delgado, H. (2021, November 30). Antiviral Gene Expression. In Encyclopedia. https://encyclopedia.pub/entry/16562
Stout-Delgado, Heather. "Antiviral Gene Expression." Encyclopedia. Web. 30 November, 2021.
Copy Citation
Influenza A virus (IAV) is a respiratory virus that alone or in combination with secondary bacterial pathogens can contribute to the development of acute pneumonia in persons >65 years of age.
antiviral
H3N2
H1N1
influenza
1. Introduction
Historically, influenza has been recognized as one of the leading causes of respiratory tract infection and contributes to seasonal epidemics. Due to antigenic variation and interspecies transmission, influenza remains a threat to global health.
Production of type I interferons (IFN) represents one of the key lines of defense against influenza. Host innate immune antiviral signaling early in response to influenza is essential to inhibit early viral replication and guide the initiation of the adaptive immune response. Infection with influenza results in the production of pathogen-associated molecular patterns (PAMPs), which are recognized by pattern recognition receptors (PRR). Several PRRs, such as Toll like receptors (TLR), NOD-like receptors (NLR), and retinoic acid inducible gene-I (RIG-I) like receptors (RLR) play a key role in the activation of the antiviral immune response to influenza. TLR 3 and 7 recognize double stranded (dsRNA) and single stranded (ssRNA) RNA, respectively and can initiate TIR domain-containing adaptor inducing IFN-β (TRIF) or myeloid differentiation factor-88 (MyD88) dependent antiviral signaling [1][2][3][4][5][6].
2. Impact of Chronological Aging on Host Responses to Mouse-Adapted H1N1 and H3N2 Strains of Influenza
2.1. Morbidity and Histological Changes in the Young and Aged Murine Adult Lung
To understand if there were age-associated differences in the host response to influenza, we infected young (3 months) or aged-adult (18–20 months) mice with mouse-adapted strains of influenza (strain: A/Puerto Rico/8/1934, PR8, H1N1) or (strain: A/Aichi/2/1968, HKx31, H3N2). In response to both strains of influenza, significantly sustained loss of weight was observed in aged adult mice when compared to young (Figure 1A,B). When compared to young, there was also a significant increase in viral titer present in aged lung in response to either H1N1 (Figure 1C) or H3N2 (Figure 1D). Histological examination of lung tissue illustrated increased cellular infiltration, with a notable increase in inflammation and intra-alveolar edema detected in aged adult lung at day 7 of infection (Figure 1E). In response to H1N1, there was increased cellular recruitment to aged adult murine lung, with detectable damage to the alveolar capillary barrier (Figure 1E). Interestingly, when compared to lung tissue collected from aged H1N1 infected mice, there was less inflammation and inflammatory damage in aged H3N2 infected lung detected at day 7 post infection (Figure 1E).
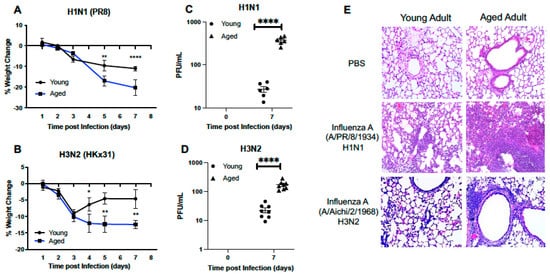
Figure 1. Morbidity and Histological Changes in the Young and Aged Murine Adult Lung. Young (3 months) and aged (18–20 months) adult mice were intranasally instilled with 12.5 PFU of influenza (strain: A/Puerto Rico/8/1934, PR8, H1N1) or (strain: A/Aichi/2/1968, HKx31, H3N2). Weight measurements were taken for young and aged mice at select time points post (A) H1N1 or (B) H3N2 infection. Viral titer in BAL was quantified and PFU/mL for (C) H1N1 and (D) H3N2. (E) Lung tissue was collected at day 7 post infection and H&E staining was performed to assess inflammation and cellular recruitment to lung. For H1N1 and H3N2 samples, each tissue section (20X magnification) shown represents a different mouse. Student’s t-test: * p < 0.05, ** p < 0.01, and **** p < 0.0001. Similar results were obtained from at least three independent experiments, with N = 10 per group. Data are expressed as the mean ± SD.
2.2. Cellular Infiltration and Lung Injury Is Increased in Aged Murine Lung in Response to Influenza
Based on these histological findings, we next examined cell numbers present in bronchoalveolar lavage (BAL) fluid isolated from young and aged adult lung at select time points post H1N1 or H3N2 infection. In response to H1N1, there was a significant increase in cells present in BAL detected at day 3 post infection (Figure 2A). Cell number continued to increase in aged lung, with significantly heightened quantities detected at day 7 post infection (Figure 2A). We next evaluated cellular numbers present in young and aged lung in response to H3N2 infection. At day 3 post infection, there was a marked increase in cellular recruitment, with significantly higher numbers of cells quantified in young lung when compared to aged (Figure 2A). By day 7 post infection, cell numbers remained significantly elevated in aged H3N2 infected lung (Figure 2A). We next examined protein levels in young and aged adult BAL samples collected from mice at day 3 and 7 post H1N1 or H3N2 infection. At day 3 post infection there was a significant increase in protein detected in aged adult lung in response to influenza, with heightened levels detected in response to H3N2 (Figure 2B). Similarly, at day 7 post influenza, there was significantly increased protein concentrations present in aged lung, with notably higher levels quantified in H3N2 lung (Figure 2B). We next investigated the amount of lung water accumulation in young and aged adult lung in response to H1N1 or H3N2 infection. At day 7 post H1N1 or H3N2 infection there was a significant increase in water accumulation in aged adult lung, as illustrated by increased wet to dry lung ratio (Figure 2C). To examine potential changes in alveolar epithelial and endothelial permeability in young and aged murine lung in response to H1N1 or H3N2, we quantified relative fluorescence levels in plasma post lung instillation with FITC. In response to influenza, there was a marked increase in permeability, as illustrated by increased FITC fluorescence in aged influenza infected lung, with significantly higher levels detected in response to H3N2 (Figure 2D).
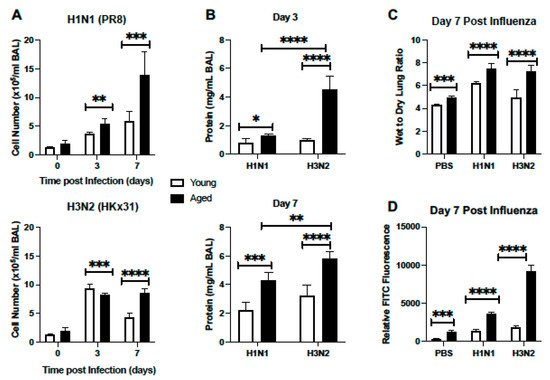
Figure 2. Cellular Infiltration and Lung Injury is Increased in Aged Murine Lung in Response to Influenza. Young (3 months) and aged (18–20 months) adult mice were intranasally instilled with 12.5 PFU of influenza (strain: A/Puerto Rico/8/1934, PR8, H1N1) or (strain: A/Aichi/2/1968, HKx31, H3N2). (A,B) At select time points post infection BAL was collected from mice and (A) cell number or (B) protein concentration was quantified. (C) Lung tissue was collected at day 7 post infection. Wet weight (initial weight upon lung tissue removal) was quantified prior to incubation at 60 °C for 48 h to yield dry weight measurements. (D) Mice were instilled on day 7 with 3mg/mL of FITC-dextran and relative FITC fluorescence in plasma was assessed. Student’s t-test: * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001. Similar results were obtained from at least three independent experiments, with N = 5 per group. Data are expressed as the mean ± SD.
2.3. Dysregulated Type I IFN Signaling in Aged Lung
Given the importance of antiviral signaling on host immune responses to influenza, we next examined the expression of several genes associated with the type I IFN signaling response in young and aged adult murine lung treated with PBS or influenza (H1N1 or H3N2) at days 3, 5, and 7 post infection. As shown in Figure 3A, there was a distinct pattern of genes elevated in both young and aged adult lung in response to each strain of influenza. On day 3, despite high Ifnα2 gene expression in aged H1N1 infected lung, we noted significantly diminished levels of IFNα present in BAL collected from H1N1 and H3N2 infected aged murine lung (Figure 3A–C). At day 3 post infection there was an increase in Ifnα2, Isg15, and Stat1 gene expression detected in young adult lung in response to either H1N1 or H3N2 (Figure 3A,C,E, and F). At day 5 of H1N1 or H3N2 infection, there was elevated expression of Ifnα2, Ifnβ1, Il-15, Mx1, and Tlr3 detected in young lung (Figure 3A,C,D). In response to H1N1 or H3N2, there was increased expression of Il-15, Tlr3, and Stat1 detected in young lung at day 7 of infection (Figure 3A,F). Examination of aged lung demonstrated differential expression patterns of genes associated with type I IFN signaling. Specifically, at day 3 post H1N1, there was increased Ifnα2 and Ifnβ1 and decreased Isg15 expression detected in aged lung (Figure 3A,C–E). In contrast, there was elevated expression of Isg15, Stat1, Mx1, and Tlr3 genes detected in aged lung at day 3 post H3N2 (Figure 3A,E,F). By days 5 and 7 of infection, there was a marked difference in gene expression with heightened expression of Mx1, Tlr3, Ifnα2, Ifnαr1, Ifnβ1, and Il-15 detected in aged lung in response to H1N1 (Figure 3A,C,D). In contrast, a similar elevation of Ifnα2, Ifnαr1, Ifnβ1, and Il-15 was not detected in aged lung in response to H3N2 (Figure 3A,C,D).
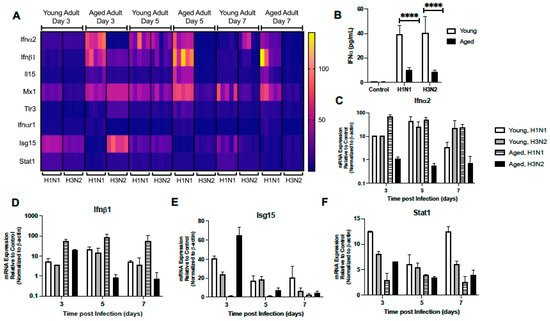
Figure 3. Dysregulated Type I IFN Signaling in Aged Lung. Young (3 months) and aged (18–20 months) adult mice were intranasally instilled with 12.5 PFU of influenza (strain: A/Puerto Rico/8/1934, PR8, H1N1) or (strain: A/Aichi/2/1968, HKx31, H3N2). (A) Lung tissue was collected from PBS treated or influenza infected young and aged mice at select time points post infection. Gene expression was assessed using the RT2 Profiler PCR Array, Mouse Antiviral Response, PAMM-122Z and results were quantified using analytical software provided by Qiagen Gene Globe. (B) IFN-α expression in BAL was assessed by ELISA on day 3 post infection (student’s t-test: **** p < 0.0001). Representative gene expression of (C) Ifnα2 (two-way ANOVA, p < 0.0001), (D) Ifnβ1 (two-way ANOVA, p = 0.0361), (E) Isg15 (two-way ANOVA, p < 0.0001), and (F) Stat1 (two-way ANOVA, p < 0.0001). Similar results were obtained from at least three independent experiments, with N = 5 per group. Data are expressed as the mean ± SD.
Table 1. Symbol and gene names.
| Symbol | Gene Name |
|---|---|
| Atg12 | Autophagy-related 12 |
| Atg5 | Autophagy-related 5 |
| Azi2 | 5-azacytidine induced gene 2 |
| Casp8 | Caspase 8 |
| Ccl3 | Chemokine (C-C motif) ligand 3 |
| Ccl4 | Chemokine (C-C motif) ligand 4 |
| Ccl5 | Chemokine (C-C motif) ligand 5 |
| Cd40 | CD40 antigen |
| Cd80 | CD80 antigen |
| Cd86 | CD86 antigen |
| Chuk | Conserved helix-loop-helix ubiquitous kinase |
| Cnpy3 | Canopy 3 homolog |
| Ctsb | Cathepsin B |
| Ctsl | Cathepsin L |
| Ctss | Cathepsin S |
| Cxcl10 | Chemokine (C-X-C motif) ligand 10 |
| Cxcl9 | Chemokine (C-X-C motif) ligand 9 |
| Cyld | Cylindromatosis |
| Ddx3x | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3 |
| Ddx58 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| Dhx58 | DEXH (Asp-Glu-X-His) box polypeptide 58 |
| Fadd | Fas (TNFRSF6)-associated via death domain |
| Ifih1 | Interferon induced with helicase C domain 1 |
| Ifnα2 | Interferon alpha 2 |
| Ifnαr1 | Interferon (alpha and beta) receptor 1 |
| Ifnβ1 | Interferon beta 1 |
| Ikbkb | Inhibitor of kappaB kinase beta |
| Il12a | Interleukin 12A |
| Il12b | Interleukin 12b |
| Il15 | Interleukin 15 |
| Il6 | Interleukin 6 |
| Irf3 | Interferon regulatory factor 3 |
| Irf7 | Interferon regulatory factor 7 |
| Isg15 | ISG15 ubiquitin-like modifier |
| Map3k1 | Mitogen-activated protein 3 kinase 1 |
| Map3k7 | Mitogen-activated protein 3 kinase 7 |
| Mapk14 | Mitogen-activated protein kinase 14 |
| Mapk8 | Mitogen-activated protein kinase 8 |
| Mavs | Mitochondrial antiviral signaling protein |
| Mx1 | Myxovirus (Influenza virus) resistance 1 |
| Nfκb1 | Nuclear factor of kappa light polypeptide gene enhancer in B- cells 1, p105 |
| Nfκbia | Nuclear factor of kappa light polypeptide gene enhancer in B- cells inhibitor, alpha |
| Pin1 | Protein (peptidyl-proyl cis/trans isomerase) NIMA-interacting 1 |
| Rela | V-rel reticuloendotheliosis viral oncogene homolog A |
| Ripk1 | Receptor (TNFRSF)-interacting serine-threonine kinase 1 |
| Stat1 | Signal transducer and activator of transcription 1 |
| Tbk1 | TANK-binding kinase 1 |
| Tlr3 | Toll-like receptor 3 |
| Tlr9 | Toll-like receptor 9 |
| Tnf | Tumor necrosis factor |
| Tradd | TNFRSF1A-associated via death domain |
| Traf3 | Tnf receptor-associated factor 3 |
| Traf6 | Tnf receptor-associated factor 6 |
| Trim25 | Tripartite motif-containing 25 |
2.4. Altered Expression of TLR Signaling Responsive Genes
We next investigated the expression pattern of TLR receptor signaling responsive genes in young and aged lung during H1N1 or H3N2 infection. In response to H1N1 or H3N2, there was a marked elevation of Ccl3, Ccl4, Ccl5, and Cxcl10 in young lung at day 3 post infection (Figure 4A–E). While some expression patterns remained elevated during infection, by day 5 there was increased Il-12a, Il-12b, Il-6, Cd80, and Cxcl11 expression that corresponded with elevated Il-15 levels in young adult H1N1 and H3N2 infected lung (Figure 4A). By day 7, expression of Ccl3, Cxcl9, Cd86, Il-6, Cd80, and Cxcl11 remained elevated in young adult lung in response to H1N1, while expression of Ccl5, Ccl4, and Cd40 remained elevated in response to either strain of influenza (Figure 4A–D). In aged adult lung, there was a significant upregulation of multiple TLR signaling responsive genes at day 3 post H1N1, with a marked increase in Cd80, Cxcl11, and Il-6 expression being detected (Figure 4A). Interestingly, by days 5 and 7, there was heightened expression of multiple genes, such as Cxcl9, Ccl3, Ccl4, Il-12a, Il-12b, Cd86, Cxcl11, and Cd80, detected in aged lung in response to H1N1 (Figure 4A–C).
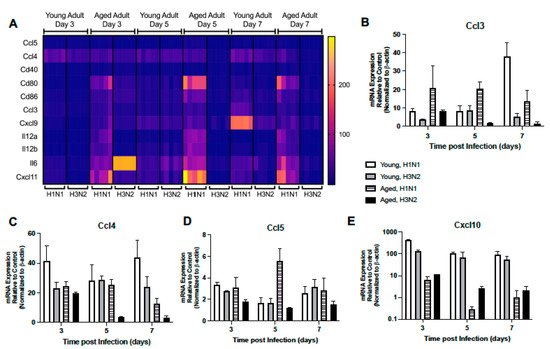
Figure 4. Altered Expression of TLR Signaling Responsive Genes. Young (3 months) and aged (18–20 months) adult mice were intranasally instilled with 12.5 PFU of influenza (strain: A/Puerto Rico/8/1934, PR8, H1N1) or (strain: A/Aichi/2/1968, HKx31, H3N2). Lung tissue was collected from PBS treated or influenza infected young and aged mice at select time points post infection. (A) Gene expression was assessed using the RT2 Profiler PCR Array, Mouse Antiviral Response, PAMM-122Z and results were quantified using analytical software provided by Qiagen Gene Globe. Representative gene expression of (B) Ccl3 (two-way ANOVA, p < 0.0001), (C) Ccl4 (two-way ANOVA, p < 0.0001), (D) Ccl5 (two-way ANOVA, p < 0.0001), and (E) Cxcl10 (p < 0.0001). Similar results were obtained from at least three independent experiments, with N = 5 per group. Data are expressed as the mean ± SD.
2.5. Altered Expression of RIG-I-like Receptor Signaling
Given the importance of RIG-I signaling in host mediated immune response to influenza, we next examined the impact of age on lung responses to H1N1 or H3N2 infection. In young lung, there was similar expression patterns in H1N1 and H3N2 lung, with increased expression of Ddx58, Trim25, and Dhx58 detected on day 3 and Cyld expression on day 5 post infection (Figure 5A–D). When compared to H3N2, by day 7, Ddx58, Dhx58, and Ifih1 expression remained elevated in young H1N1 infected lung (Figure 5A,C,E). In contrast, there was altered expression in aged lung on day 3 post infection, with increased Dhx58, Cyld, Trim25, and Ddx58 detectable in aged H1N1 infected lung tissue (Figure 5A–D). Despite increased Ifih1 gene expression on day 3 post infection, diminished RIG-I like receptor signaling was observed in aged H3N2 infected lung on day 5 and 7 post infection (Figure 5A–E).
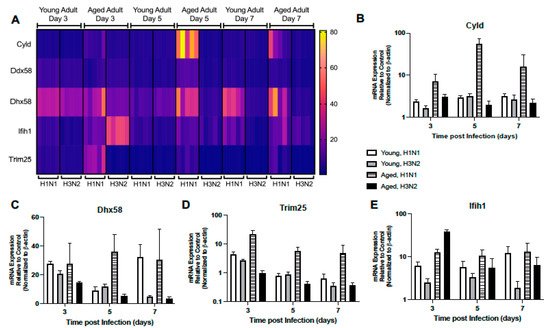
Figure 5. Altered Expression of RIG-I-Like Receptor Signaling. Young (3 months) and aged (18–20 months) adult mice were intranasally instilled with 12.5 PFU of influenza (strain: A/Puerto Rico/8/1934, PR8, H1N1) or (strain: A/Aichi/2/1968, HKx31, H3N2). Lung tissue was collected from PBS treated or influenza infected young and aged mice at select time points post infection. (A) Gene expression was assessed using the RT2 Profiler PCR Array, Mouse Antiviral Response, PAMM-122Z and results were quantified using analytical software provided by Qiagen Gene Globe. Representative expression of (B) Cyld (two-way ANOVA, p < 0.0001), (C) Dhx58 (two-way ANOVA, p = 0.0058), (D) Trim25 (two-way ANOVA, p < 0.0001), and (E) Ifih1 (two-way ANOVA, p < 0.0001). Similar results were obtained from at least three independent experiments, with N = 5 per group. Data are expressed as the mean ± SD.
We investigated the impact of aging on the expression of downstream RIG-I-like receptor signaling molecules. In young lung, there were comparable Pin1, Iκbκb, Mapk14, Map3k1, Rela, Irf7, Map3k7, and Nfκb1 expression patterns at days 3 and 5 post H1N1 or H3N2 infection (Figure 6A–C). In response to H1N1 or H3N2 infection, on day 5 post infection there were also similar expression patterns of Mavs, Traf3, and Traf6 detected in young lung (Figure 6A). Interestingly, by day 7 of infection, multiple genes were elevated in young H1N1 infected lung, such as Atg5, Mapk8, Tbk1, Casp8, and Ripk1, that were not similarly expressed in response to H3N2 (Figure 6A). In aged adult lung, there was a differential pattern of gene expression observed in response to H1N1 or H3N2 infection (Figure 6A). Specifically, at day 3, while the expression of Tbk1, Tnf, Mavs, and Traf6 were upregulated in response to H1N1, only the expression of Ddx3x, Chuk, Irf7, and Nfκbia were highly expressed in aged lung in response to H3N2 (Figure 6A,B,D). By days 5 and 7, heightened expression of Pin1, Traf6, Ripk1, Map3k7, Map3k1, Mapk14, Atg5, and TNF were observed in aged lung in response to H1N1 (Figure 6A,D).
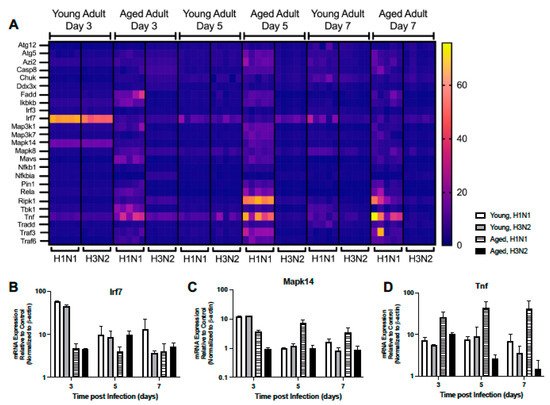
Figure 6. Altered Expression of RIG-I-Like Receptor Signaling. Young (3 months) and aged (18–20 months) adult mice were intranasally instilled with 12.5 PFU of influenza (strain: A/Puerto Rico/8/1934, PR8, H1N1) or (strain: A/Aichi/2/1968, HKx31, H3N2). Lung tissue was collected from PBS treated or influenza infected young and aged mice at select time points post infection. (A) Gene expression was assessed using the RT2 Profiler PCR Array, Mouse Antiviral Response, PAMM-122Z and results were quantified using analytical software provided by Qiagen Gene Globe. Representative expression of (B) Irf7 (two-way ANOVA, p < 0.0001), (C) Mapk14 (two-way ANOVA, p < 0.0001), and (D) Tnf (two-way ANOVA, p = 0.00459). Similar results were obtained from at least three independent experiments, with N = 5 per group. Data are expressed as the mean ± SD.
References
- Matsumoto, M.; Seya, T. TLR3: Interferon induction by double-stranded RNA including poly(I:C). Adv. Drug Deliv. Rev. 2008, 60, 805–812.
- Yamamoto, M.; Sato, S.; Hemmi, H.; Hoshino, K.; Kaisho, T.; Sanjo, H.; Takeuchi, O.; Sugiyama, M.; Okabe, M.; Takeda, K.; et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 2003, 301, 640–643.
- Oshiumi, H.; Matsumoto, M.; Funami, K.; Akazawa, T.; Seya, T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 2003, 4, 161–167.
- Sato, A.; Iwasaki, A. Induction of antiviral immunity requires Toll-like receptor signaling in both stromal and dendritic cell compartments. Proc. Natl. Acad Sci. USA 2004, 101, 16274–16279.
- Iwasaki, A.; Medzhitov, R. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 2004, 5, 987–995.
- Lund, J.M.; Alexopoulou, L.; Sato, A.; Karow, M.; Adams, N.C.; Gale, N.W.; Iwasaki, A.; Flavell, R.A. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad Sci. USA 2004, 101, 5598–5603.
More
Information
Subjects:
Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
692
Revisions:
2 times
(View History)
Update Date:
01 Dec 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




