You're using an outdated browser. Please upgrade to a modern browser for the best experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Peixin Dong | + 2564 word(s) | 2564 | 2021-11-18 08:40:22 | | | |
| 2 | Yvaine Wei | Meta information modification | 2564 | 2021-11-29 04:27:57 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Dong, P. LncRNAs: Novel Biomarkers for Pancreatic Cancer. Encyclopedia. Available online: https://encyclopedia.pub/entry/16477 (accessed on 31 December 2025).
Dong P. LncRNAs: Novel Biomarkers for Pancreatic Cancer. Encyclopedia. Available at: https://encyclopedia.pub/entry/16477. Accessed December 31, 2025.
Dong, Peixin. "LncRNAs: Novel Biomarkers for Pancreatic Cancer" Encyclopedia, https://encyclopedia.pub/entry/16477 (accessed December 31, 2025).
Dong, P. (2021, November 29). LncRNAs: Novel Biomarkers for Pancreatic Cancer. In Encyclopedia. https://encyclopedia.pub/entry/16477
Dong, Peixin. "LncRNAs: Novel Biomarkers for Pancreatic Cancer." Encyclopedia. Web. 29 November, 2021.
Copy Citation
Pancreatic cancer is one of the most deadly neoplasms and the seventh major cause of cancer-related deaths among both males and females. Long non-coding RNAs (lncRNAs) are RNAs longer than 200 nucleotides and have no protein-coding capacity. lncRNAs have been recently found to influence the progression and initiation of pancreatic cancer. MACC1-AS1, LINC00976, LINC00462, LINC01559, HOXA-AS2, LINC00152, TP73-AS1, XIST, SNHG12, LUCAT1, and UCA1 are among the oncogenic lncRNAs in pancreatic cancer.
lncRNAs
pancreatic cancer
biomarker
prognosis
non-coding RNA
microRNA
1. Oncogenic LncRNAs in Pancreatic Cancer
Expression of lncRNAs has been appraised in pancreatic cancer tissues and cell lines using lncRNA microarray and qRT-PCR methods. These methods have resulted in the identification of numerous differentially expressed lncRNAs between neoplastic and non-neoplastic tissues. For instance, MACC1-AS1 has been identified as the most over-expressed lncRNA in pancreatic cancer tissues in a study conducted by Qi C et al. [1]. Expression of MACC1-AS1 was particularly elevated in patients who had poor survival. MACC1-AS1 silencing suppresses the proliferation and metastatic ability of pancreatic cancer cells. Mechanistically, MACC1-AS1 enhances the expression of PAX8 protein, which, in turn, increases aerobic glycolysis and activates NOTCH1 signaling. In addition, the expression of PAX8 is increased in pancreatic cancer tissues in correlation with levels of MACC1-AS1 and the prognosis of patients with pancreatic cancer. Thus, the MACC1-AS1/PAX8/NOTCH1 axis has been suggested as a putative target for the treatment of pancreatic cancer [1]. The association between expression levels of LINC00976 and pancreatic cancer progression has been assessed using in situ hybridization (ISH) and qRT-PCR methods. LINC00976 is up-regulated in pancreatic cancer tissues and cell lines in correlation with poor survival of patients. LINC00976 silencing inhibits proliferation, migratory potential and invasiveness of pancreatic cancer in vivo and in vitro. LINC00976 has been found to target ovarian tumor proteases OTUD7B. This protein deubiquitinates EGFR and influences the activity of MAPK signaling. Further studies have shown the role of the LINC00976/miR-137/OTUD7B axis in the modulation of the proliferation of pancreatic cancer cells [2]. Expression of LINC00462 in pancreatic cancer cells is induced by OSM. Up-regulation of this lncRNA has been accompanied by enhancement of cell proliferation, acceleration of cell cycle progression, and inhibition of cell apoptosis and adhesion. Moreover, LINC00462 up-regulation increases the migration and invasiveness of pancreatic cancer cells through enhancement of epithelial-mesenchymal transition (EMT) and accelerated growth and metastasis of pancreatic cancer in vivo. Notably, LINC00462 has been demonstrated to have interaction with miR-665. Up-regulation of LINC00462 leads to the enhancement of expressions of TGFBR1 and TGFBR2 and the subsequent activation of the SMAD2/3 pathway in pancreatic cancer [3]. Table 1 shows the information on oncogenic lncRNAs in pancreatic cancer. Figure 1 illustrates the role of various lncRNAs in pancreatic cancer through regulating the TGF-β/SMAD signaling pathway.
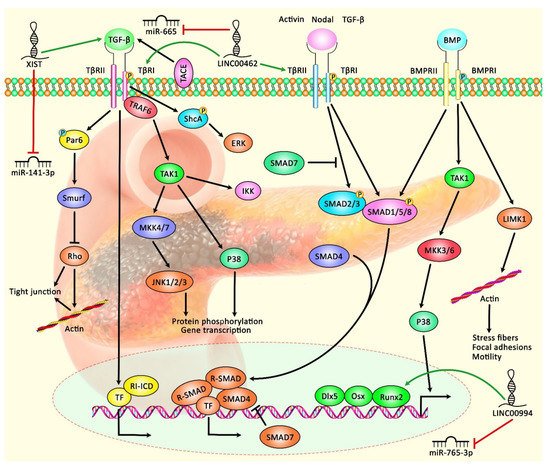
Figure 1. A schematic diagram shows the role of various lncRNAs in modulating the TGF-β/SMAD signaling pathway in pancreatic cancer. According to this cascade, when bioavailable TGF-β binds a homodimer of TβRII, transphosphorylation of the TβRI can trigger the activation of kinase activity. SMAD proteins, the substrates for TβRI kinases, are downstream of the BMP–analogous ligand–receptor systems. SMAD1, SMAD2, SMAD3, SMAD5, and SMAD8 can bind to membrane-bound serine/threonine receptors and are up-regulated via their kinase function. As a co-factor, the Co-SMAD (SMAD4) can bind to the up-regulated R-SMAD to create a complex that translocates into the nucleus. Consequently, I-SMAD (SMAD7) can deactivate the impacts of R-SMADs [4][5]. Previous studies have authenticated that several lncRNAs can play an effective role in regulating the TGF-β/SMAD cascade in pancreatic cancer. LINC00462 can up-regulate expression levels of TGFBR1 and TGFBR2 and activate the SMAD2/3 pathway in pancreatic cancer cells through down-regulating miR-665 expression [3]. Furthermore, lncRNA XIST can promote TGF-β2 expression via inhibiting the expression of miR-141-3p, thus enhancing cell proliferation, migration, and invasion of PC cells [6]. Green arrows indicate the up-regulation of target genes by lncRNAs; red arrows depict the inhibitory effects of lncRNAs.
Table 1. List of up-regulated lncRNAs in pancreatic cancer (ANT: adjacent non-cancerous tissue; cell lines were used for functional studies, apoptotic assays, and identification of partners of lncRNAs).
| LncRNA | Sample | Cell Line | Interaction | Signaling Pathway | Clinical Properties | Method | Function | Ref. |
|---|---|---|---|---|---|---|---|---|
| C9orf139 | 54 pairs of tumor and ANTs | AsPC-1, BxPC3, PANC1, PaCa-2, SW-1990, HPDE6-C7 |
miR-663a/Sox12 | _ | Tumor stage, lymph nodemetastasis | qRT-PCR, Western blotting, RNA immunoprecipitation, RNA pull-down, luciferase reporter assay |
High expression of LncRNA C9orf139 is associated with the poor clinicopathological feature of PC patients | [7] |
| CRNDE | 58 pairs of tumor and ANTs | SW-1990, PANC-1,CAPAN-1, JF305 BxPC-3,HPDE6-C7 | miR-384 | _ | Tumordifferentiation, tumor size, TNM stage, and lymph nodal metastasis | qRT-PCR, luciferase reporter assays, Western blotting, immunohistochemistry (IHC) analysis |
LncRNA CRNDE plays an oncogenic role in PC tissue and cell lines via directly targeting miR-384 | [8] |
| H19 | 139 invasive ductal carcinoma samples | PANC-1 | - | - | In situ hybridization, DNA microarray analysis, qRT-PCR | H19 affects cell motility but not cell growth rate | [9] | |
| HOTAIR | _ | HPDE6-C7, SU.86.86, CFPPAC-1, SW-1990, PL45 |
miR-34a | JAK2/STAT3 Pathway | _ | qRT-PCR, Western blotting, RNA pull-down | LncRNA HOTAIR can activate the JAK2/STAT3 pathway by targeting miR-34 and then enhancing the proliferation and invasion of PC cells | [10] |
| HOTTIP | Panc-1, L3.6pL, and MiaPaCa2 | HOXA10, HOXB2, HOXA11, HOXA9, and HOXA1 | - | - | Illumina Human V.3 HT12 Beadchip array | HOTTIP regulates the proliferation, apoptosis, and migration of PC cells | [11] | |
| HOXA-AS2 | 16 pairs of tumor and ANTs, 12 pairs of tumor and ANTs |
AsPC-1, BxPC-3, PANC-1 | enhancer of zeste homolog 2 (EZH2), lysine-specific demethylase 1 (LSD1) |
_ | _ | qRT-PCR | lncRNA HOXA-AS2 plays an oncogenic role in pancreatic cancer tissue | [12] |
| LINC00976 | _ | CFPAC-1, MIA-PaCa-2, PANC-1, BxPC-3,CFPAC-1, ASPC-1, Panc03.27, Capan-2 | miR-137/ OTUD7B |
EGFR/MAPK signaling pathway | Tumor size, lymph node metastasis, perineural invasion, vascularinvasion, distant metastasis ability |
In situ hybridization (ISH), qRT-PCR | LINC00976 plays an oncogenic role in pancreatic cancer tissue and promotes invasion, migration, and proliferation via up-regulating OTUD7B and then targeting miR-137 | [2] |
| LINC00462 | _ | SW-1990, BxPC3,PANC-1, AsPC-1, CFPAC-1, HPDE6-C7 |
miR-665, TGFBR1, TGFBR2 |
SMAD2/3 signaling pathway | Large tumor size, poor tumor differentiation, TNM stage, distant metastasis |
qRT-PCR, CCK-8 assay, Western blotting, flow cytometry analyses, immunofluorescence |
Over-expression of LINC00462 significantly promotes EMT and cell proliferation and suppresses cell apoptosis via up-regulating TGFBR1 and TGFBR2 | [3] |
| LINC01559 | 55 pairs of tumor and ANTs | AsPC-1, BxPC-3, PANC-1, MIA- PaCa-2, SW-1990, HPDE |
miR-1343-3p/RAF1 | ERK signaling pathway | Large tumors, lymph node metastasis, | RT-qPCR, RIP assay, CCK-8 assay, Western blotting, immunohistochemistry (IHC) |
High expression of LINC01559 enhances proliferation of pancreatic cancer cells and metastasis by up-regulating Raf1 and activating the ERK pathway | [13] |
| LINC00152 | 28 pairs of tumor and ANTs | BxPC3, Panc1, AsPC1, SW-1990, HPDE6-C7 | miR-150 | _ | _ | qRT-PCR, CCK-8 assay, EDU assay, luciferase reporter assay | LINC00152 can suppress miR-150 and then promote pancreatic cancer cells progressions | [14] |
| LINC00958 | _ | PANC-1, Capan-2, SW-1990, BxPC-3,HPDE | miR-330-5p | _ | _ | qRT-PCR, Western blotting, fluorescent in situ hybridization (FISH), RNA immunoprecipitation (RIP) |
LINC00958 enhances the EMT process and metastatic ability of PC cells | [15] |
| LUCAT1 | 60 pairs of tumor and ANTs | BxPC-3, Capan2, AsPC-1, PANC-1, HPDE6c7 | miR-539 | _ | tumor size, lymphatic invasion. |
qRT-PCR, in situ hybridization, Western blotting |
LUCAT1 can enhance the invasion ability of cells by targeting miR-539 | [16] |
| LINC00994 | 10 pairs of tumor and ANTs | PANC-1, AsPC-1, SW-1990 | miR-765-3p/RUNX2 | _ | _ | Microarrays, qRT-PCR, flow cytometry, luciferase assay, Western blotting |
LINC00994 acts as an oncogene and its inhibition can suppress RUNX2 by targeting miR-765-3p | [17] |
| LINC01207 | 36 pairs of tumor and ANTs | PANC-1, BxPC-3, Mpanc-96, PaTu-8988 | miR-143-5p | _ | _ | qRT-PCR, RNA pull-down, RNA immunoprecipitation (RIP), flow cytometry, immunofluorescence staining, Western blotting |
Its inhibition can induce apoptosis and autophagy activity of PC cells via targeting miR-143-5p | [18] |
| MACC1-AS1 | 98 pairs of tumor and ANTs, 124 pairs of tumor and ANTs |
BxPC-3, PANC-1, MIA-PaCa-2, KP-2, AsPC-1, Capan-1 |
PAX8 | NOTCH1 signaling pathway |
_ | lncRNA microarray, qRT-PCR, luciferase analyses, RNAimmunoprecipitation |
High expression of LncRNA MACC1-AS1 can induce pancreatic cancer cells proliferation and promote metastasis through regulating the PAX8/NOTCH1 signaling pathway | [1] |
| OIP5-AS1 | 110 pairs of tumor and ANTs | PANC-1, BxPC-3, AsPC-1, CFPAC-1, HPDE6-C7 | miR-429, FOXD1, ERK pathway |
ERK pathway | Tumor size, distant metastasis, TNM stage | qRT-PCR, RNA immunoprecipitation, RNA pull-down, luciferase reporter assay, Western blotting |
High expression of LncRNA OIP5-AS1 can increase EMT process, invasion, and PC cell proliferation via activating the ERK pathway | [19] |
| PVT1 | 30 pairs of tumor and ANTs | HPAC, DANG, BxPC-3, PANC1, ASPC-1, H6C7 | miR-519d-3p | glycolysispathway | lymph node metastasis | qRT-PCR, Western blotting, RNA immunoprecipitation (RIP) assay, RNA pull-down assay, immunohistochemistry (IHC) | PTV1 induces downregulation of miR-519d-3p and then promotes the progression of pancreatic cancer | [20] |
| RP11-567G11.1 | 78 tumor tissues and 7 non-tumor tissues | SW-1990, BxPC-3, PANC-1 | _ | NOTCH signaling pathway | _ | In situ hybridization, CCK8 and flow cytometry, Western blotting, qPCR |
Inhibition of LncRNA RP11-567G11.1 can induce apoptosis and suppress cancer cell proliferation | [21] |
| SBF2-AS1 | _ | PANC-1, BxPC-3, SW-1990, Capan2, THP-1 |
miR-122-5p | SMAD signaling pathway | _ | Flow cytometry, RNA-fluorescence in situ hybridization(FISH), qRT-PCR, Western blotting, RNA immunoprecipitation, RNA pull-down assays |
The expression level of SBF2-AS1 is increased in M2 macrophage exosomes and plays an oncogenic role in pancreatic cancer tissue | [22] |
| SNHG7 | 50 pairs of tumor and ANTs | PANC-1, SW-1990, BxPC-3 AsPC-1, HPDE | miR-146b-5, roundabout homolog 1(Robo1) |
_ | Tumor size, distant metastasis, lymph node metastasis, |
qRT-PCR, Flow cytometry analysis, luciferase reporter assay, RNA immunoprecipitation (RIP) assay, RNA pull-down assay, Western blotting | High expression of LncRNA SNHG7 can promote the progression of PC by positively affecting Robo1 | [23] |
| SNHG12 | 15 pairs of tumor and ANTs | HPDE6, BxPC-3, CAPAN1, PANC1, SW-1990 | miR-320b | _ | _ | qRT-PCR, flow cytometry, luciferase assay | LncRNA SNHG12 can increase the invasion, EMT, and proliferation of cancer cells by negatively affecting miR-320b | [24] |
| SNHG14 | 45 pairs of tumor and ANTs | CFPAC-1, BxPC-3, L3.6pl Panc-1, HPDE6C7 | miR-613 | _ | Poor tumor differentiation, advanced TNM stage, nodal metastasis | qRT-PCR, fluorescent in situ hybridization, flow cytometry, Western blotting |
Increased expression of SNHG14 can promote the progression of pancreatic cancer by inhibiting caspase-3 activity and down-regulation of miR-613 | [25] |
| SNHG15 | 48 pairs of tumor and ANTs | AsPC-1, BxPC-3, HPDE6 | zeste homolog 2 | _ | tumor size, TNM stage,lymph node, metastasis | qRT-PCR, Flow cytometry, Western blotting, RNA immunoprecipitation, chromatin immunoprecipitation (ChIP) |
SNHG15 plays an oncogenic role in pancreatic cancer tissue by inversely regulating target genes | [26] |
| SPRY4-IT1 | _ | BxPC-3, PANC-1 | Cdc20 | _ | _ | qRT-PCR, Western blotting, wound healing assay, Transwell assay | SPRY4-IT1 acts as an oncogene in PC tissue, and its inhibition induces depletion of PC progression | [27] |
| TP73-AS1 | 77 pairs of tumor and ANTs | HPDE6-C7, SW-1990, CAPAN-1, JF305, PANC-1, BxPC-3 |
miR-141 | _ | TNM stage, lymph node metastasis |
qRT-PCR, luciferase reporter assays, Western blotting | High expression of lncRNA TP73-AS1 induces migration, invasion, and PC cell proliferation | [28] |
| UCA1 | 120 pairs of tumor and ANTs | PANC-1, BxPC-3, Capan-1, SW-1990, HPDE6C-7 |
_ | _ | Tumor size, depth of invasion, CA19-9 level, tumor stage |
qRT-PCR, flow cytometry, Western blotting, |
Low expression of LncRNA UCA1 can reduce the proliferation of PC cells and induce cell cycle arrest | [29] |
| UCA1 | 36 pairs of tumor and ANTs | HPC-Y5, PANC-1, SW-1990, AsPC-1 | miR-96/FOXO3 | _ | _ | qRT-PCR, Western blotting, immunohistochemistry, flow cytometry, luciferase assay, RNA in situ hybridization |
LncRNA UCA1 acts as an oncogene in PC tissue and cell lines via negative regulating miR-96 | [30] |
| XIST | 30 pairs of tumor and ANTs | PANC-1, HEK293T | miR-141-3p, TGF-β2 | TGF-β signaling pathway | _ | qRT-PCR, luciferase reporter assay, Western blotting |
LncRNA XIST plays an oncogenic role in PC tissue through targeting miR-141-3p and the TGF-β signaling pathway | [6] |
| XIST | 64 pairs of tumor and ANTs | H6c7, Patu8988,SW-1990, BxPC-3, AsPC-1, CFPAC-1, PANC-1 |
miR-133a/EGFR | EGFR/Akt signaling | Larger tumor size, perineuralinvasion, lymph node metastasis, shorter overall survival | qRT-PCR, BrdU cell proliferation assay, luciferase reporter assay | LncRNA XIST can induce PC cell proliferation through negatively regulating miR133a and positively regulating EGFR | [31] |
| ZEB2-AS1 | 39 pairs of tumor and ANTs | AsPC-1, HPAC, Cfpac-1, PANC-1, HPDE | miR-204/ HMGB1 |
_ | _ | q-RT-PCR, Western blotting immunofluorescence assay, luciferase reporter assay, RNA-binding protein immunoprecipitation (RIP) assay, LncRNA array |
Overexpression of LncRNA ZEB2-AS1 induces cell proliferation and invasion by negatively affecting miR-204 | [32] |
2. Tumor Suppressor LncRNAs in Pancreatic Cancer
LINC01111 is a newly identified lncRNA that is significantly down-regulated in tissue and plasma samples gathered from patients with pancreatic cancer. This lncRNA has been found to exert tumor-suppressive effects. Notably, expression levels of LINC01111 have been inversely correlated with the TNM stage but positively correlated with the survival rate of patients with pancreatic cancer. LINC01111 can suppress the proliferation, cell cycle progression, invasiveness, and migratory potential of pancreatic cancer cells in vitro. Moreover, it can suppress the tumorigenic potential and metastatic ability of neoplastic cells in vivo. LINC01111 over-expression leads to the up-regulation of DUSP1 through sponging miR-3924. These events result in the inhibition of phosphorylation of SAPK, therefore inactivating SAPK/JNK signaling in pancreatic cancer cells [33]. LINC01963 is another down-regulated lncRNA in clinical samples of pancreatic cancer and cell lines. Over-expression of LINC01963 leads to suppression of colony formation, attenuation of cell cycle progression, and inhibition of proliferation and invasion of pancreatic cancer cells while enhancing the apoptosis rate in these cells. More importantly, short hairpin RNA targeting LINC01963 increases the tumorigenicity of pancreatic cancer cells in vivo. Functionally, LINC01963 decreases the expression of miR-641, a miRNA that down-regulates TMEFF2. Thus, LINC01963 suppresses the progression of pancreatic cancer through the miR-641/TMEFF2 axis [34]. MEG3 is another down-regulated lncRNA in pancreatic cancer cells, the expression of which has been inversely correlated with the expression of PI3K. In clinical samples, expression of MEG3 has been inversely correlated with tumor dimension, organ metastasis, and vascular invasion in pancreatic cancer. Functionally, MEG3 can suppress the progression of pancreatic cancer through regulation of the activity of PI3K/AKT/Bcl-2/Bax/cyclin D1/P53 and PI3K/AKT/MMP-2/MMP-9 axes [35]. On the other hand, the lncRNA GAS5 has been found to suppress metastasis of pancreatic cancer via the regulation of the miR-32-5p/PTEN axis [36].
3. Diagnostic Role of LncRNAs in Pancreatic Cancer
The diagnostic role of lncRNAs in pancreatic cancer is appraised by depicting receiver operating characteristic (ROC) curves. The calculated values for the area under these curves (AUC values) for a number of these lncRNAs are more than 0.8, suggesting the potential of these lncRNAs as diagnostic markers for pancreatic cancer (Table 2). For instance, the expression of LINC00675 is firstly assessed in a small cohort of PDAC tissues and chronic pancreatitis tissues through microarray screening. At the next step, these results are validated in larger cohorts of patients using the qRT-PCR method. Over-expression of LINC00675 is significantly correlated with lymph node metastasis, perineural invasion, and poor clinical outcome of patients with pancreatic cancer. Notably, this lncRNA has a 0.893 AUC value for predicting the progression of pancreatic cancer within one year. Moreover, the AUC value for the prediction of tumor progression within six months is 0.928. Thus, LINC00675 is a potential diagnostic marker for the prediction of recurrence in PDAC patients following radical surgical resection [37].
Table 2. Diagnostic role of lncRNAs in pancreatic cancer (ANT: adjacent non-cancerous tissue).
| LncRNA | Expression Pattern | Detection Method for LncRNAs | Sample | Area Under the Curve (AUC) | References |
|---|---|---|---|---|---|
| LncRNA-UFC1 | Up-regulation | qRT-PCR | 48 serum samples of patients | 0.810 | [38] |
| RP11-263F15.1 | Up-regulation | Microarray, qRT-PCR | 71 pairs of tumor and ANTs | 0.843 | [39] |
| ABHD11-AS1 | Up-regulation | qRT-PCR | 15 serum samples of patients and 30 healthy individuals | 0.887 | [40] |
| LINC00675 | Up-regulation | Microarray, qRT-PCR | 45 pairs of tumor and ANTs | 0.928 | [37] |
| HULC | Up-regulation | qRT-PCR | 60 serum samples of patients and 60 healthy individuals | 0.856 | [41] |
| C9orf139 | Up-regulation | qRT-PCR | 54 pairs of tumor and ANTs | 0.923 | [42] |
| PVT1 | Up-regulation | qRT-PCR | Salivary samples from 55 patients with resectable pancreatic cancer, 20 patients with benign pancreatic lesions, and 55 normal controls | 0.84 (cancer vs. benign lesion), 0.90 (cancer vs. healthy state) | [43] |
| HOTAIR | Up-regulation | qRT-PCR | 0.86 (cancer vs. benign lesion), 0.88 (cancer vs. healthy state) |
References
- Qi, C.; Xiaofeng, C.; Dongen, L.; Liang, Y.; Liping, X.; Yue, H.; Jianshuai, J. Long non-coding RNA MACC1-AS1 promoted pancreatic carcinoma progression through activation of PAX8/NOTCH1 signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 1–12.
- Lei, S.; He, Z.; Chen, T.; Guo, X.; Zeng, Z.; Shen, Y.; Jiang, J. Long noncoding RNA 00976 promotes pancreatic cancer progression through OTUD7B by sponging miR-137 involving EGFR/MAPK pathway. J. Exp. Clin. Cancer Res. 2019, 38, 1–15.
- Zhou, B.; Guo, W.; Sun, C.; Zhang, B.; Zheng, F. Linc00462 promotes pancreatic cancer invasiveness through the miR-665/TGFBR1-TGFBR2/SMAD2/3 pathway. Cell Death Dis. 2018, 9, 1–15.
- Yoshimatsu, Y.; Watabe, T. Roles of TGF-β signals in endothelial-mesenchymal transition during cardiac fibrosis. Int. J. Inflamm. 2011, 2011, 724080.
- Xu, F.; Liu, C.; Zhou, D.; Zhang, L. TGF-β/SMAD pathway and its regulation in hepatic fibrosis. J. Histochem. Cytochem. 2016, 64, 157–167.
- Sun, J.; Zhang, Y. LncRNA XIST enhanced TGF-β2 expression by targeting miR-141-3p to promote pancreatic cancer cells invasion. Biosci. Rep. 2019, 39, BSR20190332.
- Agiannitopoulos, K.; Samara, P.; Papadopoulou, M.; Efthymiadou, A.; Papadopoulou, E.; Tsaousis, G.N.; Mertzanos, G.; Babalis, D.; Lamnissou, K. miRNA polymorphisms and risk of premature coronary artery disease. Hell. J. Cardiol. 2020, 62, 278–284.
- Wang, G.; Pan, J.; Zhang, L.; Wei, Y.; Wang, C. Long non-coding RNA CRNDE sponges miR-384 to promote proliferation and metastasis of pancreatic cancer cells through upregulating IRS 1. Cell Prolif. 2017, 50, e12389.
- Yoshimura, H.; Matsuda, Y.; Yamamoto, M.; Michishita, M.; Takahashi, K.; Sasaki, N.; Ishikawa, N.; Aida, J.; Takubo, K.; Arai, T.; et al. Reduced expression of the H19 long non-coding RNA inhibits pancreatic cancer metastasis. Lab. Investig. 2018, 98, 814–824.
- Deng, S.; Wang, J.; Zhang, L.; Li, J.; Jin, Y. LncRNA HOTAIR Promotes Cancer Stem-Like Cells Properties by Sponging miR-34a to Activate the JAK2/STAT3 Pathway in Pancreatic Ductal Adenocarcinoma. OncoTargets Ther. 2021, 14, 1883.
- Cheng, Y.; Jutooru, I.; Chadalapaka, G.; Corton, J.C.; Safe, S. The long non-coding RNA HOTTIP enhances pancreatic cancer cell proliferation, survival and migration. Oncotarget 2015, 6, 10840–10852.
- Lu, J.; Wei, J.-H.; Feng, Z.-H.; Chen, Z.-H.; Wang, Y.-Q.; Huang, Y.; Fang, Y.; Liang, Y.-P.; Cen, J.-J.; Pan, Y.-H. miR-106b-5p promotes renal cell carcinoma aggressiveness and stem-cell-like phenotype by activating Wnt/β-catenin signalling. Oncotarget 2017, 8, 21461.
- Chen, X.; Wang, J.; Xie, F.; Mou, T.; Zhong, P.; Hua, H.; Liu, P.; Yang, Q. Long noncoding RNA LINC01559 promotes pancreatic cancer progression by acting as a competing endogenous RNA of miR-1343-3p to upregulate RAF1 expression. Aging 2020, 12, 14452.
- Yuan, Z.-J.; Yu, C.; Hu, X.-F.; He, Y.; Chen, P.; Ouyang, S.-X. LINC00152 promotes pancreatic cancer cell proliferation, migration and invasion via targeting miR-150. Am. J. Transl. Res. 2020, 12, 2241.
- Chen, S.; Chen, J.-Z.; Zhang, J.-Q.; Chen, H.-X.; Qiu, F.-N.; Yan, M.-L.; Tian, Y.-F.; Peng, C.-H.; Shen, B.-Y.; Chen, Y.-L. Silencing of long noncoding RNA LINC00958 prevents tumor initiation of pancreatic cancer by acting as a sponge of microRNA-330-5p to down-regulate PAX8. Cancer Lett. 2019, 446, 49–61.
- Chakraborty, T.; Bhattacharyya, A.; Pattnaik, M. Theta autoregressive neural network model for COVID-19 outbreak predictions. Medrxiv 2020.
- Zhu, X.; Niu, X.; Ge, C. Inhibition of LINC00994 represses malignant behaviors of pancreatic cancer cells: Interacting with miR-765-3p/RUNX2 axis. Cancer Biol. Ther. 2019, 20, 799–811.
- Liu, C.; Wang, J.-O.; Zhou, W.-Y.; Chang, X.-Y.; Zhang, M.-M.; Zhang, Y.; Yang, X.-H. Long non-coding RNA LINC01207 silencing suppresses AGR2 expression to facilitate autophagy and apoptosis of pancreatic cancer cells by sponging miR-143-5p. Mol. Cell. Endocrinol. 2019, 493, 110424.
- Wu, L.; Liu, Y.; Guo, C.; Shao, Y. LncRNA OIP5-AS1 promotes the malignancy of pancreatic ductal adenocarcinoma via regulating miR-429/FOXD1/ERK pathway. Cancer Cell Int. 2020, 20, 1–13.
- Sun, J.; Zhang, P.; Yin, T.; Zhang, F.; Wang, W. Upregulation of LncRNA PVT1 facilitates pancreatic ductal adenocarcinoma cell progression and glycolysis by regulating MiR-519d-3p and HIF-1A. J. Cancer 2020, 11, 2572.
- Huang, R.; Nie, W.; Yao, K.; Chou, J. Depletion of the lncRNA RP11-567G11. 1 inhibits pancreatic cancer progression. Biomed. Pharmacother. 2019, 112, 108685.
- Yin, Z.; Zhou, Y.; Ma, T.; Chen, S.; Shi, N.; Zou, Y.; Hou, B.; Zhang, C. Down-regulated lncRNA SBF2-AS1 in M2 macrophage-derived exosomes elevates miR-122-5p to restrict XIAP, thereby limiting pancreatic cancer development. J. Cell. Mol. Med. 2020, 24, 5028–5038.
- Jian, Y.; Fan, Q. Long non-coding RNA SNHG7 facilitates pancreatic cancer progression by regulating the miR-146b-5p/Robo1 axis. Exp. Ther. Med. 2021, 21, 1–13.
- Cao, W.; Zhou, G. LncRNA SNHG12 contributes proliferation, invasion and epithelial–mesenchymal transition of pancreatic cancer cells by absorbing miRNA-320b. Biosci. Rep. 2020, 40, BSR20200805.
- Deng, P.c.; Chen, W.b.; Cai, H.h.; An, Y.; Wu, X.q.; Chen, X.m.; Sun, D.l.; Yang, Y.; Shi, L.q.; Yang, Y. LncRNA SNHG14 potentiates pancreatic cancer progression via modulation of annexin A2 expression by acting as a competing endogenous RNA for miR-613. J. Cell. Mol. Med. 2019, 23, 7222–7232.
- Al-Kafaji, G.; Al-Mahroos, G.; Abdulla Al-Muhtaresh, H.; Sabry, M.A.; Abdul Razzak, R.; Salem, A.H. Circulating endothelium-enriched microRNA-126 as a potential biomarker for coronary artery disease in type 2 diabetes mellitus patients. Biomark 2017, 22, 268–278.
- Guo, W.; Zhong, K.; Wei, H.; Nie, C.; Yuan, Z. Long non-coding RNA SPRY4-IT1 promotes cell proliferation and invasion by regulation of Cdc20 in pancreatic cancer cells. PLoS ONE 2018, 13, e0193483.
- Cui, X.-P.; Wang, C.-X.; Wang, Z.-Y.; Li, J.; Tan, Y.-W.; Gu, S.-T.; Qin, C.-K. LncRNA TP73-AS1 sponges miR-141-3p to promote the migration and invasion of pancreatic cancer cells through the up-regulation of BDH2. Biosci. Rep. 2019, 39, BSR20181937.
- Chen, P.; Wan, D.; Zheng, D.; Zheng, Q.; Wu, F.; Zhi, Q. Long non-coding RNA UCA1 promotes the tumorigenesis in pancreatic cancer. Biomed. Pharmacother. 2016, 83, 1220–1226.
- Zhou, Y.; Chen, Y.; Ding, W.; Hua, Z.; Wang, L.; Zhu, Y.; Qian, H.; Dai, T. LncRNA UCA1 impacts cell proliferation, invasion, and migration of pancreatic cancer through regulating miR-96/FOXO3. Iubmb Life 2018, 70, 276–290.
- Wei, W.; Liu, Y.; Lu, Y.; Yang, B.; Tang, L. LncRNA XIST promotes pancreatic cancer proliferation through miR-133a/EGFR. J. Cell. Biochem. 2017, 118, 3349–3358.
- Gao, H.; Gong, N.; Ma, Z.; Miao, X.; Chen, J.; Cao, Y.; Zhang, G. LncRNA ZEB2-AS1 promotes pancreatic cancer cell growth and invasion through regulating the miR-204/HMGB1 axis. Int. J. Biol. Macromol. 2018, 116, 545–551.
- Pan, S.; Shen, M.; Zhou, M.; Shi, X.; He, R.; Yin, T.; Wang, M.; Guo, X.; Qin, R. Long noncoding RNA LINC01111 suppresses pancreatic cancer aggressiveness by regulating DUSP1 expression via microRNA-3924. Cell Death Dis. 2019, 10, 1–16.
- Li, K.; Han, H.; Gu, W.; Cao, C.; Zheng, P. Long non-coding RNA LINC01963 inhibits progression of pancreatic carcinoma by targeting miR-641/TMEFF2. Biomed. Pharmacother. 2020, 129, 110346.
- Gu, L.; Zhang, J.; Shi, M.; Zhan, Q.; Shen, B.; Peng, C. lncRNA MEG3 had anti-cancer effects to suppress pancreatic cancer activity. Biomed. Pharmacother. 2017, 89, 1269–1276.
- Gao, Z.-Q.; Wang, J.-f.; Chen, D.-H.; Ma, X.-S.; Wu, Y.; Tang, Z.; Dang, X.-W. Long non-coding RNA GAS5 suppresses pancreatic cancer metastasis through modulating miR-32-5p/PTEN axis. Cell Biosci. 2017, 7, 1–12.
- Li, D.-D.; Fu, Z.-Q.; Lin, Q.; Zhou, Y.; Zhou, Q.-B.; Li, Z.-H.; Tan, L.-P.; Chen, R.-F.; Liu, Y.-M. Linc00675 is a novel marker of short survival and recurrence in patients with pancreatic ductal adenocarcinoma. World J. Gastroenterol. 2015, 21, 9348.
- Liu, P.; Sun, Q.-Q.; Liu, T.-X.; Lu, K.; Zhang, N.; Zhu, Y.; Chen, M. Serum lncRNA-UFC1 as a potential biomarker for diagnosis and prognosis of pancreatic cancer. Int. J. Clin. Exp. Pathol. 2019, 12, 4125.
- Huang, X.; Ta, N.; Zhang, Y.; Gao, Y.; Hu, R.; Deng, L.; Zhang, B.; Jiang, H.; Zheng, J. Microarray analysis of the expression profile of long non-coding RNAs indicates lncRNA RP11-263F15. 1 as a biomarker for diagnosis and prognostic prediction of pancreatic ductal adenocarcinoma. J. Cancer 2017, 8, 2740.
- Liu, Y.; Feng, W.; Liu, W.; Kong, X.; Li, L.; He, J.; Wang, D.; Zhang, M.; Zhou, G.; Xu, W. Circulating lncRNA ABHD11-AS1 serves as a biomarker for early pancreatic cancer diagnosis. J. Cancer 2019, 10, 3746.
- Ou, Z.-L.; Luo, Z.; Lu, Y.-B. Long non-coding RNA HULC as a diagnostic and prognostic marker of pancreatic cancer. World J. Gastroenterol. 2019, 25, 6728.
- Ge, J.-N.; Di Yan, C.-L.G.; Wei, M.-J. LncRNA C9orf139 can regulate the growth of pancreatic cancer by mediating the miR-663a/Sox12 axis. World J. Gastrointest. Oncol. 2020, 12, 1272.
- Xie, Z.; Chen, X.; Li, J.; Guo, Y.; Li, H.; Pan, X.; Jiang, J.; Liu, H.; Wu, B. Salivary HOTAIR and PVT1 as novel biomarkers for early pancreatic cancer. Oncotarget 2016, 7, 25408–25419.
More
Information
Subjects:
Oncology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
654
Revisions:
2 times
(View History)
Update Date:
29 Nov 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




