
Video Upload Options
Emergence and rapid spread of antibiotic resistance has posed a serious threat to public health and undermined decades of progress made in the fight against bacterial infections. Plasmid-mediated horizontal tranfer of antibiotic resistance genes (ARGs) has been recognized as the most dominant dissemination pathway of ARGs in humans, animals and environmental settings. In particular, four pathways including conjugation, transformation, transduction and vesiduction account for horizontal transfer of antibiotic resistance genes. A better understanding of these pathways and underlying mechanisms would contribute to developing more effective stategies to control the prevalence of ARGs.
1. Introduction
The rapid emergence and dissemination of antibiotic resistance are increasing threats to public health [1][2]. It has been forecast that about 10 million lives would be lost due to infections caused by multidrug resistant bacteria in 2050 if the current situation continues [3]. Importantly, plasmid-mediated intra- and inter-species horizontal gene transfer (HGT) is commonly acknowledged as a major driver for the prevalence and spread of antibiotic resistance genes (ARGs) in the environment, human beings and animals [4][5]. For instance, since blaNDM-1 gene-mediated carbapenems resistance was first identified in 2009 [6], this gene has been widely reported in clinically relevant pathogens from human and animal sources [7]. Additionally, the mobilized colistin resistance gene mcr-1-positive Enterobacteriaceae [8] from different origins has been identified in over 50 countries across six continents. Meanwhile, various mcr-1 variants, such as mcr-2/3/4/5/6/7/8/9/10, were also identified in bacteria from various sources [9]. Previous research has shown that sub-minimum inhibition concentrations (MIC) of specific antibiotics can profoundly facilitate the conjugation process, thus improving the relatively low conjugation efficiency in the experimental conditions [10]. However, other compounds, such as environmental contaminants, also play a crucial role in the dissemination of ARGs, whereas their actions are largely neglected. Therefore, extensive attention should be paid to these exogenous compounds that contribute to HGT. A better understanding of the effect and molecular mechanisms of these non-antibiotic drugs on HGT would be conducive to developing more effective strategies to control the spread of ARGs. Meanwhile, the effect of these potential inhibitors of HGT is also worthy of remark as it provides a distinct pipeline in combating antibiotic resistance.
2. Horizontal Transfer of Antibiotic Resistance Genes
HGT generally refers to genetic communication between individuals of different species that can cross reproductive isolation barriers. Essentially, HGT represents the process of sharing genes or genetic material among individuals of different species, which adds new genetic variation to the recipient organisms and avoids the destructive effect caused by the gradual accumulation of point mutations. In addition, dominant traits in the process of life evolution can spread rapidly among individuals of different species, aiding them to quickly adapt to the new environment or obtain new resources [11]. Before widely found in eukaryotes, HGT was first identified in prokaryotes, which are smaller, easier to spread and highly adaptable to the environment, particularly in bacteria [12]. Because most point mutations are not beneficial or harmful to individuals, and prokaryotes lack heritable variation caused by sexual reproduction, HGT is an important means to obtain new genetic material for them. In addition to obligate endosymbiont bacteria, most bacteria quickly adapt to the new environment by obtaining genes from other species in the environment, which is often accompanied by the loss of other genes [13]. Altogether, HGT has been considered as the main driving force of prokaryotic evolution regardless of some debates.
Accordingly, conjugation, transformation, transduction and vesiduction were recognized as four major pathways for HGT in prokaryotes (particularly bacteria) (Figure 1) [14][15]. Among these four pathways, conjugation is commonly considered as the most dominant route [16]. Differently from transformation and transduction, conjugation requires cell-to-cell contact via cell surface pilus or adhesions, offering better protection from the disturbed environment and a more efficient means to transfer bacterial genes from a donor cell to a recipient cell (Figure 1A) [17]. Differently from the sex pilus, which helps the donor cell attach to the recipient cell produced by the donor cells in Gram-negative bacteria, the contact in Gram-positive bacteria relies on surface adhesins [18]. After the connection, the two cells will directly contact and form a coupling bridge by which DNA can be transferred from the donor to the recipient. Generally, this process includes two steps: (1) DNA is mobilized by mob genes-encoded relaxase proteins; (2) single-strand DNA is transported via a type IV secretion system (T4SS) [19].
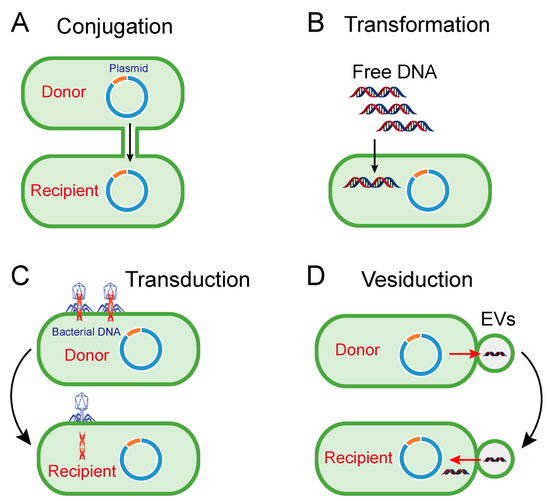
Figure 1. Pathways of horizontal gene transfer. (A) Conjugation is the process of DNA transfer from donor cell to recipient cell via cell-to cell contact. (B) Transformation represents the uptake and integration of naked fragments of extracellular DNA by recipient cells. (C,D) Transduction and vesiduction refer to the transfer of bacterial DNA (red DNA strand) by bacteriophages (C) or extracellular vesicles (EVs, D).
Although conjugation could also occur in some Gram-positive bacteria, the modes of action, particularly in cell recognition, are different from those in Gram-negative bacteria [18]. Similarly, the conjugation process in both Gram-negative and Gram-positive bacteria is highly dependent on the conjugative plasmids-encoding conjugation machinery. Meanwhile, conjugation machinery enables the transfer of non-conjugative plasmids, such as IncQ plasmids, with a broad range of hosts [20]. Notably, these mobile genetic elements, which included antibiotic resistance genes located in plasmids or chromosomes, are transferred from environment-derived bacteria to various human health-relevant pathogens [21]. Moreover, transfer of ARGs between unrelated bacteria with large taxonomic distances was also identified [22][23]. These examples suggest the remarkable role of conjugation in the dissemination of ARGs within different reservoirs.
Transformation refers to the process that extracellular free fragments of DNA are taken up and integrated by certain bacteria (Figure 1B) [24]. It requires several conditions, including free DNA fragments and competent recipient bacteria, and the translocated DNA must be integrated into the recipient genome or encircled as plasmid DNA [15]. In order to effectively absorb DNA, bacterial cells must be in a competent state, as defined by the ability of bacteria to bind to free fragments of DNA, and are formed only in a limited number of bacteria, such as Haemophilus and Streptococcus [25]. However, many other bacteria, such as E. coli, can be artificially rendered to be competent under specific stressful conditions, such as antibiotic stress or calcium chloride (CaCl2) stimulation [26]. Consistently, it has been indicated that antibiotic treatment could promote the transformation of extracellular DNA [27]. It is also noteworthy that the transformation of ARGs has been widely found in various species. For example, the fluoroquinolone resistance gene (gyrA) and penicillin resistance gene (penA) could be transformed into S. pneumoniae and commensal Neisseria species, respectively [28][29].
Unlike conjugation and transformation-mediated HGT, transduction is performed by bacteria-infecting viruses termed bacteriophages. In this process of genetic recombination, genes from host cells are first incorporated into the genome of the bacteriophage, and then carried to the recipient cell after the next bacteriophage infections (Figure 1C) [30]. Transduction is a reliable way to transfer DNA between bacteria because it can protect foreign DNA enclosed in bacteriophages from physical degradation and DNase in the environment. Additionally, cell-to-cell contact, which is indispensable in conjugation, is not necessary for transduction. However, bacteriophages-mediated transduction is not applicable for horizontal transfer of extensive genes owing to the inefficient package of bacteriophages and its high specificity to infected bacteria. Therefore, interspecific transfer caused by transduction is not common in the environmental setting. Nevertheless, several studies have suggested that bacteriophages-mediated transductions also partially account for the prevalence of ARGs. For instance, the presence of ARGs in bacteriophages has been detected in cystic fibrosis patients [31], wastewater samples [32] and animal and human fecal samples [33], suggesting that bacteriophages are potential reservoirs of ARGs.
In addition to the three canonical mechanisms above, extracellular vesicles (EVs)-mediated DNA transfer, termed vesiduction (Figure 1D), has been proposed as a fourth mode of HGT [34]. The secretion of EVs from bacterial membranes was first observed in the 1960s [35], and subsequently proven as a widespread phenomenon in both Gram-positive and Gram-negative bacteria [36][37][38][39]. Moreover, eukaryotes and archaea such as hyperthermophilic and halophilic archaea can also produce EVs [40], suggesting that this biological phenomenon is not specific to prokaryotes. It has been evidenced that EVs are associated with different physiological roles [41][42], which are highly dependent on their composition. For example, peptidoglycan hydrolases or toxin-containing EVs are involved in pathogenicity or competition with other microorganisms; quorum sensing molecules may correlate in cell-to-cell communications [38]. Additionally, EVs can function as DNA carriers capable of protecting DNA from degradation by restriction enzymes, DNase or other physical and chemical conditions, thus playing a crucial role in HGT. In 2000, Yaron et al. first indicated that EVs from E. coli O157:H7 are responsible for the transfer of virulence genes [43]. Thereafter, EVs were found to transfer endogenous and/or exogenous plasmids from cell to cell [40][44]. All examples shed light on the fact that vesiduction-conferred HGT is becoming increasingly common in diverse microorganisms. However, the precise mechanisms of vesiduction remain obscure, especially when it comes to secretion and its regulatory factors of EVs, as well as its recognition and fusion with recipient cells.
References
- Van Boeckel, T.P.; Pires, J.; Silvester, R.; Zhao, C.; Song, J.; Criscuolo, N.G.; Gilbert, M.; Bonhoeffer, S.; Laxminarayan, R. Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science 2019, 365, eaaw1944.
- Kupferschmidt, K. Resistance fighter. Science 2016, 352, 758–761.
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; 2016. (Review on Antimicrobial Resistance, London)
- Harrison, E.; Brockhurst, M.A. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 2012, 20, 262–267.
- Grohmann, E.; Muth, G.; Espinosa, M. Conjugative plasmid transfer in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2003, 67, 277–301.
- Kumarasamy, K.K.; Toleman, M.A.; Walsh, T.R.; Bagaria, J.; Butt, F.; Balakrishnan, R.; Chaudhary, U.; Doumith, M.; Giske, C.G.; Irfan, S. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010, 10, 597–602.
- Walsh, T.R.; Weeks, J.; Livermore, D.M.; Toleman, M.A. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: An environmental point prevalence study. Lancet Infect. Dis. 2011, 11, 355–362.
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168.
- Shen, Y.; Zhang, R.; Schwarz, S.; Wu, C.; Shen, J.; Walsh, T.R.; Wang, Y. Farm animals and aquaculture: Significant reservoirs of mobile colistin resistance genes. Environ. Microbiol. 2020, 22, 2469-2484.
- Andersson, D.I.; Hughes, D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478.
- Gogarten, J.P.; Doolittle, W.F.; Lawrence, J.G. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 2002, 19, 2226–2238.
- Huang, J. Horizontal gene transfer in eukaryotes: The weak-link model. Bioessays 2013, 35, 868–875.
- Ochman, H.; Lawrence, J.G.; Groisman, E.A. Lateral gene transfer and the nature of bacterial innovation. Nature 2000, 405, 299–304.
- Von Wintersdorff, C.J.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.; Wolffs, P.F. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 2016, 7, 173.
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721.
- Cabezón, E.; Ripoll-Rozada, J.; Peña, A.; de la Cruz, F.; Arechaga, I. Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 2015, 39, 81–95.
- Wozniak, R.A.; Waldor, M.K. Integrative and conjugative elements: Mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 2010, 8, 552–563.
- Kohler, V.; Keller, W.; Grohmann, E. Regulation of Gram-positive conjugation. Front. Microbiol. 2019, 10, 1134.
- Guglielmini, J.; de la Cruz, F.; Rocha, E.P. Evolution of conjugation and type IV secretion systems. Mol. Biol. Evol. 2013, 30, 315–331.
- Meyer, R. Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid 2009, 62, 57–70.
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17.
- Tamminen, M.; Virta, M.; Fani, R.; Fondi, M. Large-scale analysis of plasmid relationships through gene-sharing networks. Mol. Biol. Evol. 2012, 29, 1225–1240.
- Roberts, A.P.; Mullany, P. A modular master on the move: The Tn916 family of mobile genetic elements. Trends Microbiol. 2009, 17, 251–258.
- Domingues, S.; Harms, K.; Fricke, W.F.; Johnsen, P.J.; da Silva, G.J.; Nielsen, K.M. Natural transformation facilitates transfer of transposons, integrons and gene cassettes between bacterial species. PLoS Pathog. 2012, 8, e1002837.
- Claverys, J.P.; Martin, B.; Polard, P. The genetic transformation machinery: Composition, localization, and mechanism. FEMS Microbiol. Rev. 2009, 33, 643–656.
- Johnston, C.; Martin, B.; Fichant, G.; Polard, P.; Claverys, J.P. Bacterial transformation: Distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 2014, 12, 181–196.
- Sturød, K.; Salvadori, G.; Junges, R.; Petersen, F.C. Antibiotics alter the window of competence for natural transformation in streptococci. Mol. Oral Microbiol. 2018, 33, 378–387.
- Ferrándiz, M.J.; Fenoll, A.; Liñares, J.; De La Campa, A.G. Horizontal transfer of parC and gyrA in fluoroquinolone-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 2000, 44, 840–847.
- Bowler, L.D.; Zhang, Q.Y.; Riou, J.Y.; Spratt, B.G. Interspecies recombination between the penA genes of Neisseria meningitidis and commensal Neisseria species during the emergence of penicillin resistance in N. meningitidis: Natural events and laboratory simulation. J. Bacteriol. 1994, 176, 333–337.
- Modi, S.R.; Lee, H.H.; Spina, C.S.; Collins, J.J. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 2013, 499, 219–222.
- Rolain, J.M.; Fancello, L.; Desnues, C.; Raoult, D. Bacteriophages as vehicles of the resistome in cystic fibrosis. J. Antimicrob. Chemother. 2011, 66, 2444–2447.
- Colomer-Lluch, M.; Calero-Cáceres, W.; Jebri, S.; Hmaied, F.; Muniesa, M.; Jofre, J. Antibiotic resistance genes in bacterial and bacteriophage fractions of Tunisian and Spanish wastewaters as markers to compare the antibiotic resistance patterns in each population. Environ. Int. 2014, 73, 167–175.
- Quirós, P.; Colomer-Lluch, M.; Martínez-Castillo, A.; Miró, E.; Argente, M.; Jofre, J.; Navarro, F.; Muniesa, M. Antibiotic resistance genes in the bacteriophage DNA fraction of human fecal samples. Antimicrob. Agents Chemother. 2014, 58, 606–609.
- Soler, N.; Forterre, P. Vesiduction: The fourth way of HGT. Environ. Microbiol. 2020, 22, 2457–2460.
- Knox, K.W.; Vesk, M.; Work, E. Relation between excreted lipopolysaccharide complexes and surface structures of a lysine-limited culture of Escherichia coli. J. Bacteriol. 1966, 92, 1206–1217.
- Deatherage, B.L.; Cookson, B.T. Membrane vesicle release in bacteria, eukaryotes, and archaea: A conserved yet underappreciated aspect of microbial life. Infect. Immun. 2012, 80, 1948–1957.
- Brown, L.; Wolf, J.M.; Prados-Rosales, R.; Casadevall, A. Through the wall: Extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 2015, 13, 620–630.
- Schwechheimer, C.; Kuehn, M.J. Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat. Rev. Microbiol. 2015, 13, 605–619.
- Kim, J.H.; Lee, J.; Park, J.; Gho, Y.S. Gram-negative and Gram-positive bacterial extracellular vesicles. Semin. Cell Dev. Biol. 2015, 40, 97–104.
- Erdmann, S.; Tschitschko, B.; Zhong, L.; Raftery, M.J.; Cavicchioli, R. A plasmid from an Antarctic haloarchaeon uses specialized membrane vesicles to disseminate and infect plasmid-free cells. Nat. Microbiol. 2017, 2, 1446–1455.
- Biller, S.J.; Schubotz, F.; Roggensack, S.E.; Thompson, A.W.; Summons, R.E.; Chisholm, S.W. Bacterial vesicles in marine ecosystems. Science 2014, 343, 183–186.
- Gill, S.; Catchpole, R.; Forterre, P. Extracellular membrane vesicles in the three domains of life and beyond. FEMS Microbiol. Rev. 2019, 43, 273–303.
- Yaron, S.; Kolling, G.L.; Simon, L.; Matthews, K.R. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl. Environ. Microbiol. 2000, 66, 4414–4420.
- Gaudin, M.; Gauliard, E.; Schouten, S.; Houel-Renault, L.; Lenormand, P.; Marguet, E.; Forterre, P. Hyperthermophilic archaea produce membrane vesicles that can transfer DNA. Environ. Microbiol. Rep. 2013, 5, 109–116.




