Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | ARIF ISTIAQ | + 886 word(s) | 886 | 2021-10-08 13:44:39 | | | |
| 2 | Jessie Wu | Meta information modification | 886 | 2021-11-29 03:53:05 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Istiaq, A. Ribosome-Induced Cellular Multipotency. Encyclopedia. Available online: https://encyclopedia.pub/entry/16432 (accessed on 08 February 2026).
Istiaq A. Ribosome-Induced Cellular Multipotency. Encyclopedia. Available at: https://encyclopedia.pub/entry/16432. Accessed February 08, 2026.
Istiaq, Arif. "Ribosome-Induced Cellular Multipotency" Encyclopedia, https://encyclopedia.pub/entry/16432 (accessed February 08, 2026).
Istiaq, A. (2021, November 26). Ribosome-Induced Cellular Multipotency. In Encyclopedia. https://encyclopedia.pub/entry/16432
Istiaq, Arif. "Ribosome-Induced Cellular Multipotency." Encyclopedia. Web. 26 November, 2021.
Copy Citation
Ribosomes are generally acknowledged to perform a critical part in the translation process by decoding messenger RNA into protein. In contrast, ribosomal proteins appear to serve non-translational roles in growth, differentiation, and disease. Dr. Ohta's team recently discovered that ribosomes can revert cellular potency to a multipotent state. Ribosomal incorporation has the potential to influence the fate of both somatic and cancer cells towards multipotency, allowing them to switch cell lineage. Multipotency acquisition occurs alongside cellular stress and cell-cycle arrest in this process.
ribosome
induced pluripotency
reprogramming
1. Introduction
Ribosomes are macromolecular complexes made of proteins and RNA that serve as translation machines in the cell. Ribosomal proteins, in addition to translation, also play roles in development, differentiation, and cancer, which are referred to as “extra-ribosomal functions” [1][2]. Dr. Ohta's Lab recently demonstrated that exogenous ribosomal incorporation (the uptake of the free ribosome by living cells) into somatic and cancer cells causes the reversal of cell fate into a multipotent state [3][4][5][6]. The functional role of the ribosome as an in vitro de-differentiating factor is a recent discovery in the field of cell reprogramming; however, the molecular mechanism underlying ribosome-mediated multipotency is unknown. At the molecular level, somatic and cancer cells differ in a variety of ways. Cancer cells are less specialized than somatic cells, with uncontrolled proliferation, mutation, and altered epigenetics [7][8]. However, both somatic and cancer cells undergo cell-cycle arrest, sphere formation, and multipotency when subjected to ribosome incorporation, suggesting that a similar mechanism is at work. Such an effect on the cells has the potential to be therapeutic. Inducing cell-cycle arrest and reprogramming cancer cells to become non-cancerous is a promising cancer therapy goal [9].
2. Ribosome Incorporation for the Generation of Multipotent Cells from Somatic Cells
2.1. Ribosomes Are the Bacteria-Derived Factors Inducing Cluster Formation and Reprogramming
Endosymbiotic relationships led to the evolution of eukaryotic cells from prokaryotic organisms, and many eukaryotic organelles arose from the engulfment of previously free-living prokaryotes that were close to bacteria [10]. The symbiotic relationship between prokaryotes and eukaryotes coexisted with the evolution of simple eukaryotes into complex animals. Dr. Ohta's group previously demonstrated that lactic acid bacteria (LAB) can reprogram somatic cells, proving that bacteria can influence eukaryotic cell fate [11][12]. When they infected human dermal fibroblast cells (HDFs) with LAB in vitro, the cells (HDFs) accumulated and formed sphere-like shapes. However, the capacity of bacteria to induce sphere formation was dependent on a mild trypsinization step, as trypsinization increases membrane permeability. After 14 days of culture, sphere cells expressed pluripotency markers. They were able to differentiate into three germ-layer-derived cells, both in vitro and in vivo. Unlike iPSCs, these sphere cells expressed a subset of pluripotent markers, including NANOG, SOX2, OCT3/4, and TDGF1, but not GDF3, FGF4, REX1, or ECAT15. Another feature of these cells is their inability to proliferate, which is linked to the expression of senescent markers p15, p16, and ARF. Electron microscopy analysis of the sphere cells revealed that LAB were localized in the cytoplasm, implying that LAB in the culture found a mechanism to enter cells and induce multipotency. In a subsequent study, they revealed that the multipotency was caused by LAB ribosomes [3].To identify the responsible factor, they fractionated the LAB cell lysate using column chromatography and evaluated each fraction for sphere-inducing ability on HDFs. Sphere formation is a well-established characteristic of stem cells [13][14].Using mass spectrometry analysis, they found that the fraction with the highest activity contained ribosomal proteins. They used purified ribosomes from various sources (laboratory-generated and commercially available) and verified their ability to induce spheres, successfully establishing the ribosome as the responsible factor. In short, they introduced the exogenous ribosomes directly into the serum-free culture media containing trypsinized cells, and subsequently, the cells ingested these ribosomes by endocytosis.
These spheres (RICs, ribosome-incorporated cell clusters) formed within one to two days of ribosome incorporation in HDFs. Exogenous ribosomes were found in the cytoplasm and nucleus of RICs, as detected by immune fluorescence analysis using antibodies specific to exogenous ribosome. They used tetra-(His)6-tagged ribosomes from E. coli JE28 bacteria (a generous gift from Dr. Ederth) for this purpose, which allowed them to trace the localization of exogenous ribosomes in HDFs. Genetically modified E. coli JE28 bacteria produce functioning ribosomes with a His-tag in the L12 ribosomal protein [15]. A subset of the RICs cells expressed pluripotency markers NANOG, POU5F1 (OCT4), SOX2, and SSEA4. NANOG and OCT4 expression levels in RICs were lower than those in iPSCs. The gene expression pattern of RICs from day 0 to day 14 differed from HDFs, iPSCs, and ES cells. In the differentiation media, the RICs were differentiated into three germ-layer-derived cells. RICs also differentiated into adipocytes, osteoblasts, and chondrocytes. Figure 1 RICs, however, we're unable to differentiate in vivo, unlike bacteria-induced spheres. The characteristics of RICs place them in an unusual category of multipotent cells.
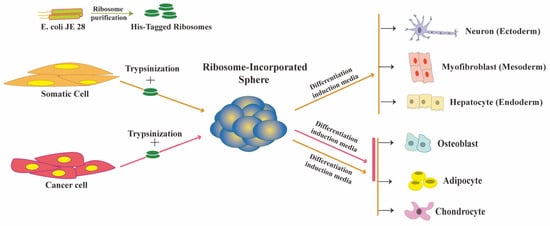
Figure 1. Ribosome incorporation leads to multipotency in somatic and cancer cells.
Surprisingly, ribosomes from various prokaryotic (Gram-positive and Gram-negative bacteria) and eukaryotic (yeast, mouse, and human) sources showed the sphere-forming effect on HDFs, suggesting a shared ribosomal element is involved in the mechanism. Although the composition and structure of ribosomes varies between prokaryotes and eukaryotes, thirty-four ribosomal proteins are conserved (15 small subunit proteins and 19 large subunit proteins) between them [16]. These proteins, however, are not fully conserved; therefore, the reprogramming function of the ribosome may be linked to sequence similarity.
References
- Warner, J.R.; McIntosh, K.B. How Common Are Extra-Ribosomal Functions of Ribosomal Proteins? Mol. Cell 2009, 34, 3–11.
- Zhou, X.; Liao, W.-J.; Liao, J.-M.; Liao, P.; Lu, H. Ribosomal Proteins: Functions beyond the Ribosome. J. Mol. Cell Biol. 2015, 7, 92–104.
- Ito, N.; Katoh, K.; Kushige, H.; Saito, Y.; Umemoto, T.; Matsuzaki, Y.; Kiyonari, H.; Kobayashi, D.; Soga, M.; Era, T.; et al. Ribosome Incorporation into Somatic Cells Promotes Lineage Transdifferentiation towards Multipotency. Sci. Rep. 2018, 8, 1634.
- Ito, N.; Anam, M.B.; Ahmad, S.A.I.; Ohta, K. Transdifferentiation of Human Somatic Cells by Ribosome. Dev. Growth Differ. 2018, 60, 241–247.
- Anam, M.B.; Istiaq, A.; Kariya, R.; Kudo, M.; Ishtiyaq Ahmad, S.A.; Ito, N.; Okada, S.; Ohta, K. Ribosome Induces Transdifferentiation of A549 and H-111-TC Cancer Cell Lines. Biochem. Biophys. Rep. 2021, 26, 100946.
- Kudo, M.; Anam, M.B.; Istiaq, A.; Ahmad, S.A.I.; Ito, N.; Ohta, K. Ribosome Incorporation Induces EMT-like Phenomenon with Cell Cycle Arrest in Human Breast Cancer Cell. Cells Tissues Organs 2021, 1–10.
- Lodish, H.; Berk, A.; Zipursky, S.L.; Matsudaira, P.; Baltimore, D.; Darnell, J. Tumor Cells and the Onset of Cancer. In Molecular Cell Biology, 4th ed.; Macmillan: New York, NY, USA, 2000.
- Interplay between the Cancer Genome and Epigenome: Cell. Available online: https://www.cell.com/fulltext/S0092-8674(13)00296-1 (accessed on 23 March 2021).
- Otto, T.; Sicinski, P. Cell Cycle Proteins as Promising Targets in Cancer Therapy. Nat. Rev. Cancer 2017, 17, 93–115.
- Cornish-Bowden, A. Lynn Margulis and the Origin of the Eukaryotes. J. Theor. Biol. 2017, 434, 1.
- Ohta, K.; Kawano, R.; Ito, N. Lactic Acid Bacteria Convert Human Fibroblasts to Multipotent Cells. PLoS ONE 2012, 7, e51866.
- Ito, N.; Ohta, K. Reprogramming of Human Somatic Cells by Bacteria. Dev. Growth Differ. 2015, 57, 305–312.
- Pastrana, E.; Silva-Vargas, V.; Doetsch, F. Eyes Wide Open: A Critical Review of Sphere-Formation as an Assay for Stem Cells. Cell Stem Cell 2011, 8, 486–498.
- Reynolds, B.A.; Weiss, S. Generation of Neurons and Astrocytes from Isolated Cells of the Adult Mammalian Central Nervous System. Science 1992, 255, 1707–1710.
- Ederth, J.; Mandava, C.S.; Dasgupta, S.; Sanyal, S. A Single-Step Method for Purification of Active His-Tagged Ribosomes from a Genetically Engineered Escherichia Coli. Nucleic Acids Res. 2009, 37, e15.
- Lecompte, O.; Ripp, R.; Thierry, J.; Moras, D.; Poch, O. Comparative Analysis of Ribosomal Proteins in Complete Genomes: An Example of Reductive Evolution at the Domain Scale. Nucleic Acids Res. 2002, 30, 5382–5390.
More
Information
Subjects:
Developmental Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
773
Revisions:
2 times
(View History)
Update Date:
29 Nov 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




