Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Hui Teng Tan | + 1951 word(s) | 1951 | 2021-11-10 04:59:37 | | | |
| 2 | Catherine Yang | Meta information modification | 1951 | 2021-11-19 09:49:06 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Tan, H.T. Phycobiliproteins. Encyclopedia. Available online: https://encyclopedia.pub/entry/16184 (accessed on 08 February 2026).
Tan HT. Phycobiliproteins. Encyclopedia. Available at: https://encyclopedia.pub/entry/16184. Accessed February 08, 2026.
Tan, Hui Teng. "Phycobiliproteins" Encyclopedia, https://encyclopedia.pub/entry/16184 (accessed February 08, 2026).
Tan, H.T. (2021, November 19). Phycobiliproteins. In Encyclopedia. https://encyclopedia.pub/entry/16184
Tan, Hui Teng. "Phycobiliproteins." Encyclopedia. Web. 19 November, 2021.
Copy Citation
Phycobiliproteins are gaining popularity as long term, high value natural products which can be alternative to synthetic products. This study analyzed research trends of phycobiliproteins from 1909 to 2020 using a bibliometric approach based on the Scopus database.
microalgae
phycobiliproteins
bibliometric
pigment
applications
1. Research Trend in Phycobiliproteins Research
Keywords are the basis of bibliographic research of academic literature [1]. Keyword analysis indicates researchers’ emphasis on a specific study topic, making it an important component of bibliometric analysis [2]. The visualization network map (Figure 1) was created with VOSviewer to assess the occurrence relationships between the keywords collected from phycobiliprotein research articles [3]. Five clusters with different colors were determined in the keyword map, with a high degree of overlap.
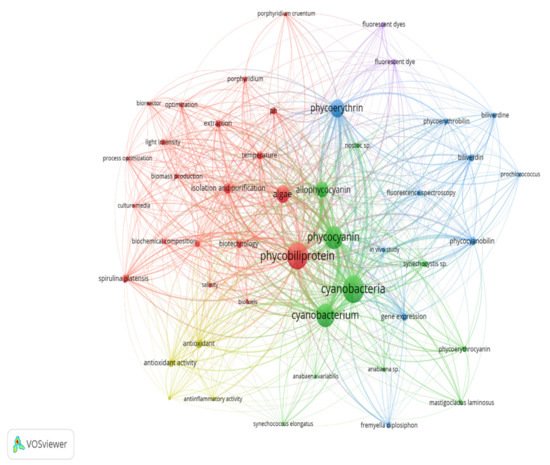
Figure 1. The bibliometric map based on total co-occurrence analysis with network visualization from 1909 to 2020 (minimum occurrences: 20).
1.1. Optimization of Cyanobacteria Cultivation and Phycobiliproteins Harvesting Process
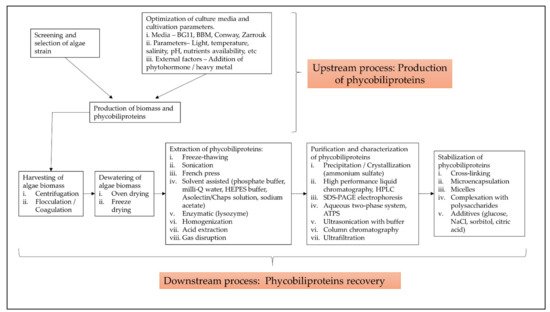
The term “phycobiliprotein” was grouped with other 20 terms in cluster 1 (Figure 1, in red), which focuses primarily on process optimization of algae cultivation, algae biomass harvesting, and phycobiliprotein production. Successful cultivation technologies rely on algae species. In addition, phycobiliprotein production is also affected by cultural conditions and nutrition factors. These factors must be optimized to develop a feasible, sustainable, and economically viable culture system for algae [4][5]. The production of high-purity phycobiliproteins comprised a series of concomitant steps that included two main sequential processes, upstream and downstream processes, as shown in Figure 2. For high-quality phycobiliproteins, the optimum production conditions and parameters are required. Hence, it is critical to focus on each step of the phycobiliproteins production process to improve the accumulation of high-quality phycobiliproteins from each species [6]. Several studies have been carried out in depth for this purpose [6][7][8][9]. For example, Manirafasha et al. [10] reviewed the techniques to increase phycobiliprotein production, from algae strain selection to culture parameter optimization and phycobiliprotein extraction to phycobiliprotein purification. Furthermore, Begum et al. [11] discussed the effect of different drying methods on the production and purity of phycobiliproteins. In addition, Lo et al. [12] reviewed the procedure followed to increase phycocyanin and phycoerythrin production yield and stability.
1.2. Classification with Structure
The term “cyanobacteria” was the central theme of the keyword map. It is part of cluster 2 (Figure 1, in green), which included 11 additional keywords. The classification of phycobiliproteins was the focus of this cluster. Phycobiliproteins are classified primarily by their absorbance spectrum properties: phycocyanin, PC (Amax = about 620 nm), phycoerythrin, PE (Amax = about 560 nm), and allophycocyanin, APC (Amax = about 650 nm) [6]. Each phycobiliprotein comprises a different polypeptide subunit (α and ß), containing covalently linked open-chain tetrapyrrole chromophores [15]. The chromophores, known as phycobilins, are covalently linked to the proteins via one or two thioether bonds to specific cysteine residues [16]. There are various structurally distinct phycobilin chromophores having distinctive spectroscopic characteristics that are also modulated by the phycobiliprotein quaternary structure [6]. The chromophores phycourobilin (PUB), phycobiliviolin (PXB), phycoerythrobilin (PEB), and phycocyanobilin (PCB) exhibit different absorbance maxima at around 498 nm, 568 nm, 535 to 567 nm, and 620 to 660 nm, respectively, when covalently linked to phycobiliproteins [16]. Furthermore, phycobiliprotein that exist as aggregates of heterodimers of alpha and beta subunits have several highly conserved amino acid residues necessary for αß heterodimer formation and chromophore binding [17]. Hence, it should be expected that some novel phycobiliproteins will be discovered throughout time. For example, Montgomery et al. [18] discovered a new and unique set of proteins that are most closely linked to allophycocyanin members of the phycobiliprotein superfamily. Each of these proteins are known as allophycocyanin-like (Apl) proteins. Novel chromophore types have been discovered in Cryptomonad phycobiliproteins [19][20]. The phycobiliproteins are further classified into many subtypes based on the chromophores’ number, combination, and position [19][20].
1.3. Gene Expression of the Phycobiliproteins Biosynthesis Pathway
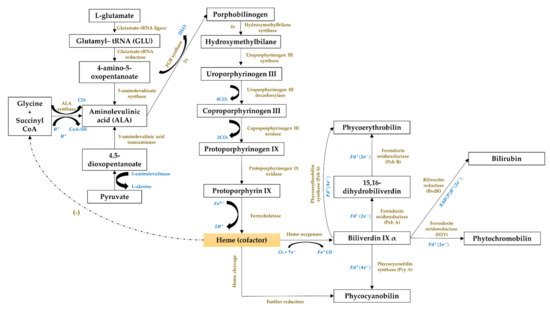
The third cluster (Figure 1, in blue) focused on the gene expression of the phycobiliproteins biosynthesis pathway. The biosynthesis of phycobiliproteins occurs via transcription, translation, and posttranslational pathways, which included the synthesis of amino acids, proteins, and phycobilins, as well as the ligation of phycobilins formed to apoproteins during the posttranslational phase (Figure 3) [21][22]. Heme acts as a cofactor in various biological roles, including enzyme activity regulation, and represents the appropriate metabolic path of all bilin biosynthesis pathways [23]. Biliverdin is a common intermediate and precursor in the biosynthesis of bilins [24]. However, biliverdin sometimes will not be an intermediate if the reduction process happens before heme cleavages [24]. Manirafasha et al. [10] and Pagels et al. [6] summarized the mechanism of phycobiliprotein biosynthesis.
1.4. Bioactivities and Applications of Phycobiliproteins
The theme of the fourth cluster (Figure 1, in yellow) was phycobiliprotein bioactivities. Natural sources of bioactive compounds are gaining popularity, as they can benefit humans [27]. The rising demand for natural bioactive molecules extracted using a simple approach has sparked the attention of researchers to explore unusual sources, including cyanobacteria [6]. Phycobiliproteins are highly valued natural products with various applications, including medicinal, nutraceutical, food, feed, and cosmetics. The research on phycobiliproteins bioactivities has grown in recent years, yet most of the research was focused on the bioactivities of phycocyanin [28][29][30]. For instance, Fernández-Rojas et al. [31] and Yu et al. [13] reviewed the bioactivities of phycocyanin. Furthermore, Pagels et al. [6] described the bioactivities from phycobiliproteins such as antioxidant capacity, anticancer, anti-inflammatory, anti-diabetes, antibacterial, anti-obesity, and neuroprotector agent. The fifth cluster (Figure 6, in purple) focused on the phycobiliprotein applications especially as the fluorescent dye.
2. Future Research Prospects in Phycobiliproteins Study
Research frontier refers to a growing trend in research theory and subject content that may be represented using burst keywords [32]. Kleinberg introduced the burst detection approach in 2002. Burst keywords are terms that suddenly increase within a short time [33]. It is possible to reveal the information that does not meet the frequency criteria but has informatics importance in academic advancement using the burst detection approach. It may be more practical and scientific to depict interaction and development trend of research frontiers by identifying hotspots change [34]. CiteSpace (a type of citation visualization software) was used to create the scientific knowledge mapping of burst detection to assess the research hotspots of phycobiliproteins [35][36]. Over time, the research emphases and orientations can be more directly represented by analyzing the changes in the most used author keywords in different periods. This study demonstrated that the earlier publications related to phycobiliprotein were its role in photosynthesis and energy transfer to chlorophyll, resulting in a citation burst of “energy transfer” and “photosynthesis” from 1976 [37][13][38]. Phycoerythrin was widely explored due to its fluorescent properties [39][40]. Furthermore, the application of phycoerythrin and allophycocyanin in flow cytometry has gained prominence since 1986 [41][42]. The majority of the phycobiliproteins were identified in cyanobacteria, especially Arthrospira platensis. The bioactivity properties of phycobiliproteins (specifically antioxidant activity) have piqued the interest of researchers since 2011 and have remained a research frontier until now [6][10]. Economically sustainable and environmentally friendly phycobiliprotein extraction methods have also gained popularity and have helped broaden the consumer acceptability of cheaper and safer natural pigments [8][9][43][44].
Phycobiliproteins are mainly found in blue-green algae, yet could also be found in red algae, cyanelles, and cryptomonads [6]. Most researchers used blue-green algae for their phycobiliproteins research, whereas only two red algae genera out of ten were exploited. The recent studies on red algae investigated the structure of phycobilisome in Griffithsia pacifica and the structural basis of energy transfer in phycobilisome of Porphyridium purpureum [45][46]. The Arhtrospira genus was the most used model organism. The Arthrospira genus is broadly recognized for its high phycocyanin content [10]. In addition, Arthrospira maxima has been commercially utilized as a food since 1521 [47]. The oldest records of production of Arthrospira biomass for human consumption are from the Aztecs [48]. Furthermore, Arthrospira has also been exploited as a protein supplement by the Kanembu tribe of Africa near Lake Chad since 1940 [49]. Synechococcus is an unicellular and euryhaline cyanobacterium [50]. Synechococcus is the most plentiful (up to 105 mL−1) and widespread picophytoplankton genus in the open ocean [51]. It is commonly used as a model organism to study cyanobacterial metabolism, particularly photosynthetic research, and has the potential for biotechnological uses [51][52]. It is also touted as a phycocyanin and phycoerythrin-rich genus [53]. Furthermore, it has a fast growth rate and an extraordinary resistance to high light irradiation [50]. Hence, these characteristics of Synechococcus favored most researchers in selecting this cyanobacterium for their study. On the other hand, Synechocystis and Nostoc genus are the other blue-green algae frequently employed in phycobiliproteins research due to their high nutritional values, and they are widely commercialized. Synechocystis has received attention in modeling studies and biotechnological applications due to a variety of characteristics including its fast growth, the potential to fix carbon dioxide into valuable products, and the relative simplicity of genetic modification [54]. Despite Synechocystis, the Nostoc genus is employed as a food and feed supplement in Mongolia, China, and South America [49]. Nostoc commune has long been recognized as a worldwide nutritious meal and traditional medicine [55]. A wide variety of notably pharmacological and protective physiological properties of the Nostoc genus aroused the attention of researchers [55]. On the other hand, the number of algae commonly claimed as toxic genera (Microcystis, Anabaena, Phormidium, and Nostoc) was lower than the nontoxic algae genera (Porphyridium, Oscillatoria, Gracilaria, Synechocystis, Arthrospira, and Synechococcus). This indicated that more studies were focused on the benefits of cyanobacteria and their bioactivities. Microcystis and Anabaena are the most important toxic cyanobacteria bloom genera in terms of diversity, impact potential, and cascading ecological effects [56][57]. Although numerous microalgae species are available in various culture collections worldwide, only a minority have been thoroughly studied [58]. Strains such as Haematococcus pluvialis (main source of astaxanthin), Dunaliella salina (the major source of beta-carotene), and Spirulina platensis (prime source of phycocyanin), are the examples of microalgae that have finally reached commercial-scale success [59][28][60]. Hundreds of many strains have been described in the literature as sources of phycobiliproteins. However, the lack of strain robustness or low productivity under outdoor environments has been typically cited as the cause of the failure of these strains in achieving commercial-scale production [58].
3. Challenges and Approaches in the Phycobiliprotein Field
The corpus of phycobiliprotein studies has been steadily enhanced and deepened due to the passion and efforts of researchers in studying phycobiliproteins. It gradually evolved from a fundamental and unitary topic to a multiperspective and sustainable development study field involving biology, chemistry, technology, and the environment. The market of phycobiliproteins will most likely continue to develop due to the rising natural product demand, the discovery of novel phycobiliproteins, advancements in the upstream and downstream processes, and expanding of the market potential [10][37]. The present study postulates that phycobiliprotein research would continue to be active and expand in bioactivity properties and applications. To meet the demand of the market, several strategies should be adopted (Figure 4). First, worldwide collaboration should be prioritized in order to conduct higher quality research. Second, most of the phycobiliproteins research is performed on a laboratory scale currently. Until now, China has achieved 200 tons/year production capacity, but the production is only limited to phycocyanin (https://www.binmei-global.com/about-us/ Assessed date: 28 June 2021). To obtain higher efficiency of growth and desired biomolecules, it is crucial to optimize relevant parameters in large-scale practical applications and develop optimal conditions of the photobioreactor [61][62]. Third, the major drawback in the use of phycobiliproteins is the high production cost. Additional screening of indigenous and novel cyanobacterial species/strains for high content of phycobiliproteins and fast-growing capability, effective harvesting, and economical purification methods may lower the production cost of phycobiliprotein. Another important aspect of phycobiliproteins research that needs to be highlighted is the taxonomy of algae species and the classification of phycobiliproteins. The use of an “omics” (genomics, metabolomics, proteomics, and transcriptomics) approach in research can enable the identification of species, detection of nutritional strategies, the interaction of symbiotic relationships with bacteria, and the biosynthesis of biomolecules, especially secondary metabolites [63]. These subject matters can help understand the metabolic pathways and regulatory mechanisms triggering the production of phycobiliproteins [61]. Besides, the development and utilization of robust platforms (Dictionary of Natural Products, MassBank, Global Natural Products Social Molecular Networking, and AntiBase) for data exchange and knowledge transfer/dissemination will undoubtedly facilitate new scientific knowledge and know-how [63].
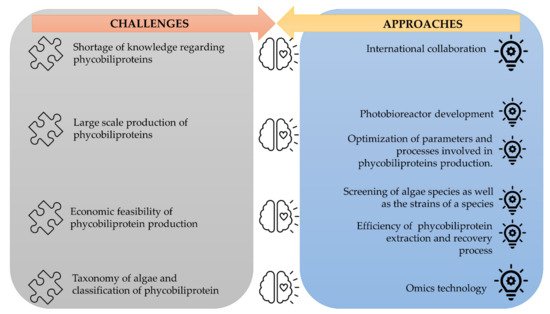
Figure 4. The challenges of phycobiliproteins research and its approaches.
References
- Jones, K.S.; Jackson, D.M. The use of automatically-obtained keyword classifications for information retrieval. Inf. Storage Retr. 1970, 5, 175–201.
- Wang, C.C.; Ho, Y.S. Research trend of metal–Organic frameworks: A bibliometric analysis. Scientometrics 2016, 109, 481–513.
- Wong, S.L.; Nyakuma, B.B.; Nordin, A.H.; Lee, C.T.; Ngadi, N.; Wong, K.Y.; Oladokun, O. Uncovering the dynamics in global carbon dioxide utilization research: A bibliometric analysis (1995–2019). Environ. Sci. Pollut. Res. 2021, 28, 13842–13860.
- Mutanda, T.; Naidoo, D.; Bwapwa, J.K.; Anandraj, A. Biotechnological applications of microalgal oleaginous compounds: Current trends on microalgal bioprocessing of products. Front. Energy Res. 2020, 8, 598803.
- Khan, M.I.; Shin, J.H.; Kim, J.D. The promising future of microalgae: Current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products. Microb. Cell Fact. 2018, 17, 36.
- Pagels, F.; Guedes, A.C.; Amaro, H.M.; Kijjoa, A.; Vasconcelos, V. Phycobiliproteins from Cyanobacteria: Chemistry and biotechnological applications. Biotechnol. Adv. 2019, 37, 422–443.
- Viskari, P.J.; Colyer, C.L. Rapid extraction of phycobiliproteins from cultured Cyanobacteria samples. Anal. Biochem. 2003, 319, 263–271.
- Tan, H.T.; Khong, N.M.H.; Khaw, Y.S.; Ahmad, S.A.; Yusoff, F.M. Optimization of the freezing-thawing method for extracting phycobiliproteins from Arthrospira sp. Molecules 2020, 25, 3894.
- Chamorro-Cevallos, G. Methods for extraction, isolation and purification of C-phycocyanin: 50 years of research in review. Int. J. Food Nutr. Sci. 2016, 3, 1–10.
- Manirafasha, E.; Ndikubwimana, T.; Zeng, X.; Lu, Y.; Jing, K. Phycobiliprotein: Potential microalgae derived pharmaceutical and biological reagent. Biochem. Eng. J. 2016, 109, 282–296.
- Begum, H.; Khatoon, H.; Yusoff, F.M.; Shariff, M. Production and purity of phycobiliproteins from selected marine and freshwater cyanobacteria subjected to different drying methods. Asian Fish. Sci. 2020, 33, 258–265.
- Hsieh-Lo, M.; Castillo, G.; Ochoa-Becerra, M.A.; Mojica, L. Phycocyanin and phycoerythrin: Strategies to improve production yield and chemical stability. Algal Res. 2019, 42, 101600.
- Yu, P.; Wu, Y.; Wang, G.; Jia, T.; Zhang, Y. Purification and bioactivities of phycocyanin. Crit. Rev. Food Sci. Nutr. 2017, 57, 3840–3849.
- Hashimoto, E.; Yabuta, Y.; Watanabe, F.; Morimoto, M.; Yamaguchi, Y.; Takenaka, H. Purification and characterization of phycobiliproteins from edible Cyanobacterium Nostochopsis sp. Food Sci. Technol. Res. 2012, 18, 485–490.
- Stadnichuk, I.N.; Tropin, I.V. Phycobiliproteins: Structure, functions and biotechnological applications. Appl. Biochem. Microbiol. 2017, 53, 1–10.
- Apt, K.E.; Collier, J.L.; Grossman, A.R. Evolution of the phycobiliproteins. J. Mol. Biol. 1995, 248, 79–96.
- Scheer, H.; Zhao, K.H. Biliprotein maturation: The chromophore attachment. Mol. Microbiol. 2008, 68, 263–276.
- Montgomery, B.L.; Casey, E.S.; Grossman, A.R.; Kehoe, D.M. AplA, a member of a new class of phycobiliproteins lacking a traditional role in photosynthetic light harvesting. Society 2004, 186, 7420–7428.
- Wedemayer, G.J.; Wemmer, D.E.; Glazer, A.N. Phycobilins of Cryptophycean algae: Structures of novel bilins with acryloyl substituents from phycoerythrin 566. J. Biol. Chem. 1991, 266, 4731–4741.
- Wedemayer, G.J.; Kidd, D.G.; Wemmer, D.E.; Glazer, A.N. Phycobilins of Cryptophycean algae. Occurrence of dihydrobiliverdin and mesobiliverdin in cryptomonad biliproteins. J. Biol. Chem. 1992, 267, 7315–7331.
- Schluchter, W.M.; Shen, G.; Alvey, R.M.; Biswas, A.; Saunée, N.A.; Williams, S.R.; Mille, C.A.; Bryant, D.A. Phycobiliprotein Biosynthesis in Cyanobacteria: Structure and Function of Enzymes Involved in Post-Translational Modification; Springer: New York, NY, USA, 2010; Volume 675.
- Belford, H.S.; Offier, G.D.; Troxler, R.F. Phycobiliprotein synthesis in the unicellular rhodophyte, Cyanidiurn cazdurium. J. Biol. Chem. 1983, 258, 4503–4510.
- McErlean, M.; Liu, X.; Cui, Z.; Gust, B.; Van Lanen, S.G. Identification and characterization of enzymes involved in the biosynthesis of pyrimidine nucleoside antibiotics. Nat. Prod. Rep. 2021, 7.
- Brown, S.B.; Holroyd, J.A.; Vernon, D.I. Biosynthesis of phycobiliproteins. Biochem. J. 1984, 219, 905–909.
- Kumar, V.; Maurya, P.K.; Mondal, S.; Sinha, R.P.; Singh, S.P. Photomorphogenesis in the Cyanobacterium Fremyella Diplosiphon Improves Photosynthetic Efficiency; Elsevier Inc.: Amsterdam, The Netherlands, 2018.
- Singh, N.K.; Sonani, R.R.; Prasad Rastogi, R.; Madamwar, D. The phycobilisomes: An early requisite for efficient photosynthesis in Cyanobacteria. EXCLI J. 2015, 14, 268–289.
- Eriksen, N.T. Production of phycocyanin—A pigment with applications in biology, biotechnology, foods and medicine. Appl. Microbiol. Biotechnol. 2008, 80, 1–14.
- Romay, C.; Gonzalez, R.; Ledon, N.; Remirez, D.; Rimbau, V. C-phycocyanin: A biliprotein with antioxidant, anti-inflammatory and neuroprotective effects. Curr. Protein Pept. Sci. 2003, 4, 207–216.
- Abd El-Baky, H.H.; El-Baroty, G.S. Characterization and bioactivity of phycocyanin isolated from Spirulina Maxima grown under salt stress. Food Funct. 2012, 3, 381–388.
- Safari, R.; Amiri, R.Z.; Esmaeilzadeh Kenari, R. Antioxidant and antibacterial activities of C-Phycocyanin from common name Spirulina platensis. Iran. J. Fish. Sci. 2020, 19, 1911–1927.
- Fernández-Rojas, B.; Hernández-Juárez, J.; Pedraza-Chaverri, J. Nutraceutical properties of phycocyanin. J. Funct. Foods 2014, 11, 375–392.
- Chen, C. Searching for intellectual turning points: Progressive knowledge domain visualization. Proc. Natl. Acad. Sci. USA 2004, 101, 5303–5310.
- Chen, C. CiteSpace II: Detecting and visualizing emerging trends and transient patterns in scientific literature. J. Assoc. Inf. Sci. Technol. 2006, 57, 359–377.
- Qi, Y.; Chen, X.; Hu, Z.; Song, C.; Cui, Y. Bibliometric analysis of algal-bacterial symbiosis in wastewater treatment. Int. J. Environ. Res. Public Health 2019, 16, 1077.
- Jiang, M.; Qi, Y.; Liu, H.; Chen, Y. The role of nanomaterials and nanotechnologies in wastewater treatment: A bibliometric analysis. Nanoscale Res. Lett. 2018, 13, 233.
- Chen, C. CiteSpace: A Practical Guide for Mapping Scientific Literature; Nova Science Publishers: Hauppauge, NY, USA, 2016.
- Sekar, S.; Chandramohan, M. Phycobiliproteins as a commodity: Trends in applied research, patents and commercialization. J. Appl. Phycol. 2008, 20, 113–136.
- Sundström, V.; van Grondelle, R. Dynamics of excitation energy transfer in photosynthetic bacteria. Chlorophylls 1991, 2137, 627–704.
- Kronick, M.N.; Grossman, P.D. Immunoassay techniques with fluorescent phycobiliprotein conjugates. Clin. Chem. 1983, 29, 1582–1586.
- Sun, L.; Wang, S.; Chen, L.; Gong, X. Promising fluorescent probes from phycobiliproteins. IEEE J. Sel. Top. Quantum Electron. 2003, 9, 177–188.
- Jung, T.M.; Dailey, M.O. A novel and inexpensive source of allophycocyanin for multicolor flow cytometry. J. Immunol. Methods 1989, 121, 9–18.
- Glazer, A.N. Phycobiliproteins—A family of valuable, widely used fluorophores. J. Appl. Phycol. 1994, 6, 105–112.
- Tavanandi, H.A.; Mittal, R.; Chandrasekhar, J.; Raghavarao, K.S.M.S. Simple and efficient method for extraction of C-phycocyanin from dry biomass of Arthospira platensis. Algal Res. 2018, 31, 239–251.
- Ruiz-Domínguez, M.C.; Jáuregui, M.; Medina, E.; Jaime, C.; Cerezal, P. Rapid green extractions of C-phycocyanin from Arthrospira Maxima for functional applications. Appl. Sci. 2019, 9, 1987.
- Zhang, J.; Ma, J.; Liu, D.; Qin, S.; Sun, S.; Zhao, J.; Sui, S.F. Structure of phycobilisome from the red alga Griffithsia pacifica. Nature 2017, 551, 57–63.
- Ma, J.; You, X.; Sun, S.; Wang, X.; Qin, S.; Sui, S.F. Structural basis of energy transfer in Porphyridium Purpureum phycobilisome. Nature 2020, 579, 146–151.
- Farrar, W.V. Tecuitlatl: A Glimpse of Aztec food technology. Nature 1966, 211, 341–342.
- Hamed, I. The evolution and versatility of microalgal biotechnology: A review. Compr. Rev. Food Sci. Food Saf. 2016, 15, 1104–1123.
- Abdulqader, G.; Barsanti, L.; Tredici, M.R. Harvest of Arthrospira Platensis from Lake Kossorom (Chad) and its household usage among the Kanembu. J. Appl. Phycol. 2000, 12, 493–498.
- Ludwig, M.; Bryant, D.A. Synechococcus Sp. Strain PCC 7002 transcriptome: Acclimation to temperature, salinity, oxidative stress, and mixotrophic growth conditions. Front. Microbiol. 2012, 3, 354.
- Coutinho, F.; Tschoeke, D.A.; Thompson, F.; Thompson, C. Comparative genomics of Synechococcus and proposal of the new GENUS Parasynechococcus. PeerJ 2016, 2016, 1–18.
- Kumar, J.; Singh, D.; Tyagi, M.B.; Kumar, A. Cyanobacteria: Applications in Biotechnology; Elsevier Inc.: Amsterdam, The Netherlands, 2019; Volume 7421.
- Wang, K.; Wommack, K.E.; Chen, F. Abundance and distribution of Synechococcus spp. and cyanophages in the Chesapeake Bay. Appl. Environ. Microbiol. 2011, 77, 7459–7468.
- Cordara, A.; Re, A.; Pagliano, C.; Van Alphen, P.; Pirone, R.; Saracco, G.; dos Santos, F.B.; Hellingwerf, K.; Vasile, N. Analysis of the light intensity dependence of the growth of Synechocystis and of the light distribution in a photobioreactor energized by 635 Nm light. PeerJ 2018, 2018, e5256.
- Li, Z.; Guo, M. Healthy efficacy of Nostoc Commune vaucher. Oncotarget 2018, 9, 14669–14679.
- Cheung, M.Y.; Liang, S.; Lee, J. Toxin-producing Cyanobacteria in freshwater: A review of the problems, impact on drinking water safety, and efforts for protecting public health. J. Microbiol. 2013, 51, 1–10.
- Saad, A.; Atia, A. Review on freshwater blue-green algae (Cyanobacteria): Occurrence, classification and toxicology. Biosci. Biotechnol. Res. Asia 2014, 11, 1319–1325.
- Garrido-Cardenas, J.A.; Manzano-Agugliaro, F.; Acien-Fernandez, F.G.; Molina-Grima, E. Microalgae research worldwide. Algal Res. 2018, 35, 50–60.
- Silva, S.C.; Ferreira, I.C.; Dias, M.M.; Barreiro, M.F. Microalgae-derived pigments: A 10-year bibliometric review and industry and market trend analysis. Molecules 2020, 25, 3406.
- Xu, Y.; Ibrahim, I.M.; Wosu, C.I.; Ben-Amotz, A.; Harvey, P.J. Potential of new isolates of Dunaliella Salina for natural β-carotene production. Biology 2018, 7, 14.
- García, J.L.; de Vicente, M.; Galán, B. Microalgae, old sustainable food and fashion nutraceuticals. Microb. Biotechnol. 2017, 10, 1017–1024.
- Yusoff, F.M.; Nagao, N.; Imaizumi, Y.; Toda, T. Bioreactor for microalgal cultivation systems: Strategy and development. In Prospects of Renewable Bioprocessing in Future Energy Systems, Biofuel and Biorefinery Technologies; Springer: Berlin/Heidelberg, Germany, 2019; pp. 117–159.
- Oliveira, C.Y.B.; Oliveira, C.D.L.; Müller, M.N.; Santos, E.P.; Dantas, D.M.M.; Gálvez, A.O. A scientometric overview of global dinoflagellate research. Publications 2020, 8, 52.
More
Information
Subjects:
Plant Sciences
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.3K
Revisions:
2 times
(View History)
Update Date:
19 Nov 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No