Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Walter Andrés Rincón Riveros | + 1380 word(s) | 1380 | 2021-10-25 10:17:19 | | | |
| 2 | Catherine Yang | Meta information modification | 1380 | 2021-11-12 02:22:01 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Rincón Riveros, W.A. NcRNAs with Regulatory Functions. Encyclopedia. Available online: https://encyclopedia.pub/entry/15921 (accessed on 07 February 2026).
Rincón Riveros WA. NcRNAs with Regulatory Functions. Encyclopedia. Available at: https://encyclopedia.pub/entry/15921. Accessed February 07, 2026.
Rincón Riveros, Walter Andrés. "NcRNAs with Regulatory Functions" Encyclopedia, https://encyclopedia.pub/entry/15921 (accessed February 07, 2026).
Rincón Riveros, W.A. (2021, November 11). NcRNAs with Regulatory Functions. In Encyclopedia. https://encyclopedia.pub/entry/15921
Rincón Riveros, Walter Andrés. "NcRNAs with Regulatory Functions." Encyclopedia. Web. 11 November, 2021.
Copy Citation
Noncoding RNAs (ncRNAs) play prominent roles in the regulation of gene expression via their interactions with other biological molecules such as proteins and nucleic acids.
genomics
transcriptome
ncRNA
lncRNA
interactome
1. Introduction
Noncoding RNA (ncRNA) genes are those genes that produce transcripts or functional RNAs that, unlike messenger RNAs, are not translated into proteins. Only a few years ago these transcripts were considered “dark matter” in the genome, but now they play leading roles in the regulation of biological processes [1][2].
In terms of their functions, ncRNAs are mainly classified (Figure 1) into housekeeping and regulatory noncoding RNAs [3][4][5][6][7][8].
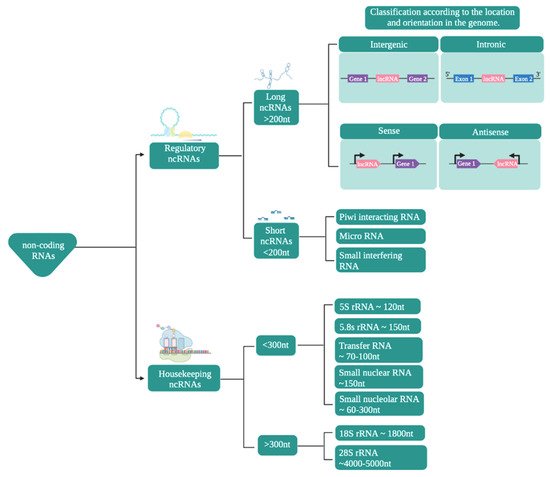
Figure 1. Classification of noncoding RNAs. The scheme presents the classification of noncoding RNAs according to their function, size, and location/orientation in the genome.
Regulatory ncRNAs (Figure 1) are classified according to their size. ncRNAs of less than 200 nucleotides are known as small ncRNAs, while those larger than that are known as long noncoding RNAs (lncRNAs) [9][10][11][12].
The small ncRNAs include microRNAs (miRNAs), small interfering RNAs (siRNAs), and Piwi-interacting RNAs (piRNAs), among which miRNAs have become particularly prominent in research as they play important roles in the regulation of numerous cellular processes, making them potential treatment targets or biomarkers [13][14][15]. Mature miRNAs are between 19 and 23 nucleotides long and are transcribed by RNA polymerase II [8][16]. siRNAs, with a size of between 18 and 30 nucleotides, are the most diverse members of this group and can be of endogenous or exogenous origin. They can also regulate expression at the transcriptional and post-transcriptional levels [17][18].
lncRNAs are defined as transcripts that are at least 200 nucleotides in length [19][20]. They share some characteristics with mRNAs, such as both having exons and introns (although lncRNAs have fewer than mRNAs), most being generated by RNA Pol II, as well as a large proportion of them have a polyadenylated tail at their 3′ end and a methylated cap at their 5′ end [21][22]. These are located in intergenic regions or between exons [2][23] and have the ability to fold in on themselves and adopt various secondary and tertiary structures that maintain similar functions in evolutionarily distant species [24]. Regarding the expression levels of lncRNAs, they are very low, with variable expression patterns depending on the tissue, stage of development, or physiological or pathological state [25][26].
2. Regulation of the Gene Expression of ncRNAs through Their Interactions with Other Biological Molecules
Many different mechanisms that regulate gene expression (Figure 2), such as transcription factors accessing DNA, and variations in the rates of mRNA synthesis, processing, stability, and translation, are influenced by ncRNAs [21][27]. This is achieved by ncRNAs’ ability to interact with various biological molecules (Figure 3) within different cells and tissues. Among these ncRNAs with regulatory functions, this review particularly focuses on miRNAs and lncRNAs, which have attracted great interest given their roles in various biological functions [13][28][29][30].
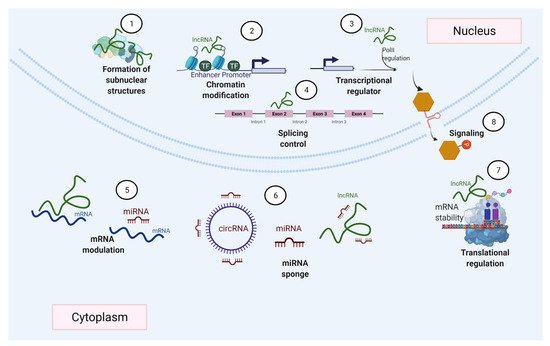
Figure 2. ncRNAs can regulate gene expression by diverse mechanisms. ncRNAs participate in the formation of nuclear bodies (1), gene transcription (2–3), modulate splicing events (4), regulate mRNA by degradation or stabilization (5), act as miRNA sponges (6), and ncRNAs can also be involved in the control of transcription (7) and cell signaling (8).
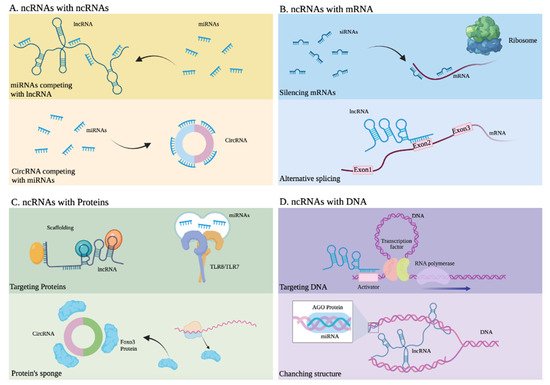
Figure 3. Interactions between noncoding RNAs and other molecules. (A) ncRNAs with other ncRNAs. Upper: miRNAs competing with lncRNA. Lower: CircRNA competing with miRNAs. (B) ncRNAs with mRNA. Upper: siRNAs silencing mRNA. Lower: Alternative splicing of mRNA due to an lncRNA. (C) ncRNAs with proteins. Upper: An lncRNA developing scaffold function and miRNAs activating Toll-like receptors. Lower: A circRNA serving as a sponge or Foxo3 protein. (D) ncRNAs with DNA. Upper: An lncRNA targeting the activator of a gene. Lower: An lncRNA altering the structure of DNA.
miRNAs are generated in the nucleus as pre-miRNAs and processed and exported to the cytoplasm by exportin 5, where they can regulate gene expression mainly at the post-transcriptional level (Figure 3A,C) by binding with other RNAs for regulatory purposes [31][32][33]. Through this mechanism, miRNAs can decrease the expression of certain proteins, through sequence complementarity with their corresponding messenger RNA or through interaction with regulatory lncRNAs. This type of regulation, mainly associated with translational repression, is the mechanism reported in most studies [34][35], along with the regulation of mRNAs through their interaction with lncRNAs. A well-documented example of a regulatory mechanism mediated by miRNAs, which function as modulator of transcripts of coding genes or their regulatory lncRNAs, involves PTEN, a tumor-suppressor gene under complex regulatory control by ncRNA [36][37]. The presence of mature miRNAs within the nucleus has also been reported; these miRNAs activate or silence genes through various mechanisms and, as a result of their direct interaction with DNA or through protein scaffolds (Figure 3D), mechanisms that include epigenetic pathways [38][39][40].
lncRNAs are characterized by their wide functional versatility because they promote the regulation of gene expression either in the nucleus or in the cytoplasm and at different levels transcriptionally and post-transcriptionally, thanks to their ability to interact with other nucleic acids such as DNA and RNA, as well as proteins (Figure 3) [41][42]. In the nucleus, they regulate gene expression in various ways. These include epigenetic modifications by directly binding to DNA and the recruitment of chromatin modifiers. This can lead to a change in the accessibility of genes to DNA-binding proteins, such as transcription factors and even RNA Pol II (Figure 3D), resulting in the activation or suppression of transcription [43][44][45][46]. Another of the most widely studied regulatory mechanisms of lncRNAs involves them acting like enhancers, in which they function either by directly interacting with promoter regions of the genes they regulate or by binding to proteins that participate as mediators [47][48][49][50][51]. These lncRNAs, through their interactions with proteins, recruit such proteins to participate in DNA repair [52]. In the nucleus, these lncRNAs can interact with mRNAs to stabilize them or to direct splicing (Figure 3B) towards a specific mRNA isoform [53][54][55]. Meanwhile, in the cytoplasm, lncRNAs display equally versatile functions. They interact with other ncRNAs such as miRNAs, or with mRNAs or proteins, through mechanisms that can result in the suppression or promotion of the products of the genes that they regulate. For example, some lncRNAs and circular RNAs regulate the activity of miRNAs because they have binding sites that retain them, thus modulating the activity of miRNAs. LncRNAs, which present this mechanism, are considered miRNA sponges, and they are part of a complex interaction network in the transcriptome or also called the theory of competitive endogenous RNAs. Then, miRNA sponges are considered exogenous when they are artificially introduced into a biological system or endogenous when they are expressed naturally [56][57][58]. In addition, ncRNAs, through various interactions with other biological molecules, can be key participants in signaling pathways [59][60][61][62][63][64].
3. The Importance of Prediction Models That Can Later Be Tested Experimentally
Recently developed laboratory-based techniques have generated major advances in the study of the interactions of RNAs with other biomolecules. These include RAP-RNA, RIA-Seq, hiCLIP, CLASH, PARIS, SPLASH, and LIGR-seq. However, the availability of these approaches is limited, and they are expensive, which impedes study of the ncRNA interactome [65][66].
By taking advantage of the information that is generated experimentally using bioinformatics and mathematical algorithms, and storing it in databases and applying open-source tools, it is possible to predict interactions between molecules such as DNA, RNA, and proteins, which can save operating costs and time (Figure 4) [67][68].
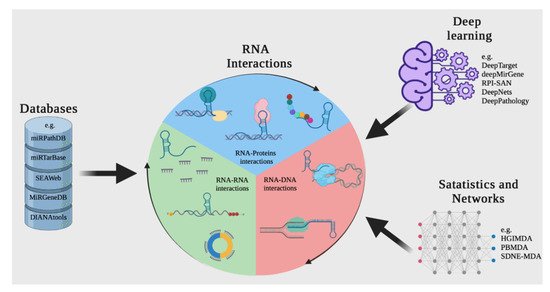
Figure 4. A computational approach for discovering or predicting RNA interactions among different biomolecules. The first strategy is to search in web tools or RNA databases such as miRTarBase. Another way to discover RNA interactions is to use resources based on deep learning and other machine learning-based tools, such as DeepTarget and deepMirGene. Finally, different mathematical and network theory methods can be used to research RNA interactions.
The advantage of using ncRNA databases and bioinformatic tools is that, thanks to the fact that they are continually fed new information obtained experimentally and supported by an exhaustive data curation process, they can support the design of experimental trials for targeting and discovering new interactions [69][70].
The various species of RNAs and their products interact in complex ways, the understanding of which provides deeper insights into the functioning of living organisms including humans. This article is intended to boost our understanding of health and disease through clarifying the intricate networks of interactions between heterogeneous biological molecules. Towards this goal, we review the latest advances in databases, bioinformatic tools, and new strategies in silico that allow the establishment or prediction of biological interactions between ncRNAs, particularly miRNAs and lncRNAs. However, these tools also have contained biomolecule-related information in another species.
References
- Zhang, Z.; Wu, H.; Peng, Q.; Xie, Z.; Chen, F.; Ma, Y.; Zhang, Y.; Zhou, Y.; Yang, J.; Chen, C.; et al. Integration of Molecular Inflammatory Interactome Analyses Reveals Dynamics of Circulating Cytokines and Extracellular Vesicle Long Non-Coding RNAs and mRNAs in Heroin Addicts During Acute and Protracted Withdrawal. Front Immunol. 2021, 12, 730300.
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 924–933.
- Natsidis, P.; Schiffer, P.H.; Salvador-Martínez, I.; Telford, M.J. Computational discovery of hidden breaks in 28S ribosomal RNAs across eukaryotes and consequences for RNA Integrity Numbers. Sci. Rep. 2019, 9, 1944.
- Krahn, N.; Fischer, J.T.; Söll, D. Naturally Occurring tRNAs With Non-canonical Structures. Front. Microbiol. 2020, 11, 596914.
- Hari, R.; Parthasarathy, S. Prediction of Coding and Non-Coding RNA; Ranganathan, S., Gribskov, M., Nakai, K., Schönbach, C.B.T.-E., Eds.; Academic Press: Oxford, UK, 2019; pp. 230–240. ISBN 978-0-12-811432-2.
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651.
- Losko, M.; Kotlinowski, J.; Jura, J. Long Noncoding RNAs in Metabolic Syndrome Related Disorders. Mediat. Inflamm. 2016, 2016, 1–12.
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, biology and functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17.
- Dahariya, S.; Paddibhatla, I.; Kumar, S.; Raghuwanshi, S.; Pallepati, A.; Gutti, R.K. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol. Immunol. 2019, 112, 82–92.
- Li, C.H.; Chen, Y. Small and Long Non-Coding RNAs: Novel Targets in Perspective Cancer Therapy. Curr. Genom. 2015, 16, 319–326.
- Sikora, M.; Marycz, K.; Smieszek, A. Small and Long Non-coding RNAs as Functional Regulators of Bone Homeostasis, Acting Alone or Cooperatively. Mol. Ther. Nucleic Acids 2020, 21, 792–803.
- Laurent, G.S.; Wahlestedt, C.; Kapranov, P. The Landscape of long noncoding RNA classification. Trends Genet. 2015, 31, 239–251.
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276.
- Tribolet, L.; Kerr, E.; Cowled, C.; Bean, A.G.D.; Stewart, C.R.; Dearnley, M.; Farr, R.J. MicroRNA Biomarkers for Infectious Diseases: From Basic Research to Biosensing. Front. Microbiol. 2020, 11, 1197.
- Galvão-Lima, L.J.; Morais, A.H.F.; Valentim, R.A.M.; Barreto, E.J.S.S. miRNAs as biomarkers for early cancer detection and their application in the development of new diagnostic tools. Biomed. Eng. Online 2021, 20, 1–20.
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402.
- Piatek, M.J.; Werner, A. Endogenous siRNAs: Regulators of internal affairs. Biochem. Soc. Trans. 2014, 42, 1174–1179.
- Claycomb, J.M. Ancient Endo-siRNA Pathways Reveal New Tricks. Curr. Biol. 2014, 24, R703–R715.
- Kazimierczyk, M.; Kasprowicz, M.K.; Kasprzyk, M.E.; Wrzesinski, J. Human Long Noncoding RNA Interactome: Detection, Characterization and Function. Int. J. Mol. Sci. 2020, 21, 1027.
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407.
- Sun, Q.; Hao, Q.; Prasanth, K.V. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet. 2018, 34, 142–157.
- Bunch, H. Gene regulation of mammalian long non-coding RNA. Mol. Genet. Genom. 2018, 293, 1–15.
- Latgé, G.; Poulet, C.; Bours, V.; Josse, C.; Jerusalem, G. Natural Antisense Transcripts: Molecular Mechanisms and Implica-tions in Breast Cancers. Int. J. Mol. Sci. 2018, 19, 123.
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nat. Cell Biol. 2009, 458, 223–227.
- Kornienko, A.E.; Dotter, C.P.; Guenzl, P.M.; Gisslinger, H.; Gisslinger, B.; Cleary, C.; Kralovics, R.; Pauler, F.M.; Barlow, D.P. Long non-coding RNAs display higher natural expression variation than protein-coding genes in healthy humans. Genome Biol. 2016, 17, 14.
- Lv, D.; Xu, K.; Jin, X.; Li, J.; Shi, Y.; Zhang, M.; Jin, X.; Li, Y.; Xu, J.; Li, X. LncSpA: LncRNA Spatial Atlas of Expression across Normal and Cancer Tissues. Cancer Res. 2020, 80, 2067–2071.
- Wei, J.-W.; Huang, K.; Yang, C.; Kang, C.-S. Non-coding RNAs as regulators in epigenetics. Oncol. Rep. 2016, 37, 3–9.
- Vidigal, J.A.; Ventura, A. The biological functions of miRNAs: Lessons from in vivo studies. Trends Cell Biol. 2015, 25, 137–147.
- Kazimierczyk, M.; Wrzesinski, J. Long Non-Coding RNA Epigenetics. Int. J. Mol. Sci. 2021, 22, 6166.
- Zhao, Z.; Sun, W.; Guo, Z.; Zhang, J.; Yu, H.; Liu, B. Mechanisms of lncRNA/microRNA interactions in angiogenesis. Life Sci. 2020, 254, 116900.
- De Sousa, M.C.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249.
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027.
- Ghafouri-Fard, S.; Bahroudi, Z.; Shoorei, H.; Abak, A.; Ahin, M.; Taheri, M. microRNA-140: A miRNA with diverse roles in human diseases. Biomed. Pharmacother. 2021, 135, 111256.
- Carrasco, I.D.; Rasco, A.G.; Fernández, A.O. ¿Qué son los micro-RNA? ¿Para qué sirven? ¿Qué potenciales beneficios podrían tener en el contexto asistencial? Cardiocore 2016, 51, 161–166.
- Chen, Y.-W.; Du, Q.-R.; He, Y.-J.; Chen, W.-S.; Jiang, W.-Y.; Gui, Q.; Xu, C.-C.; Wang, W.; Cheng, H.-Y. Circ_0044516 Regulates miR-136/MAT2A Pathway to Facilitate Lung Cancer Development. J. Immunol. Res. 2021, 2021, 5510869.
- Lou, W.; Ding, B.; Fu, P. Pseudogene-Derived lncRNAs and Their miRNA Sponging Mechanism in Human Cancer. Front. Cell Dev. Biol. 2020, 8, 85.
- Travis, G.; Haddadi, N.; Simpson, A.M.; Marsh, D.J.; McGowan, E.M.; Nassif, N.T. Studying the Oncosuppressive Functions of PTENP1PTENP1a s a ceRNA. In Pseudogenes: Functions and Protocols; Poliseno, L., Ed.; Springer: New York, NY, USA, 2021; pp. 165–185.
- Broughton, J.; Lovci, M.T.; Huang, J.L.; Yeo, E.; Pasquinelli, A.E. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol. Cell 2016, 64, 320–333.
- Dharap, A.; Pokrzywa, C.; Murali, S.; Pandi, G.; Vemuganti, R. MicroRNA miR-324-3p Induces Promoter-Mediated Expres-sion of RelA Gene. PLoS ONE 2013, 8, e79467.
- Zhou, M.; Yin, X.; Zheng, L.; Fu, Y.; Wang, Y.; Cui, Z.; Gao, Z.; Wang, X.; Huang, T.; Jia, J.; et al. miR-181d/RBP2/NF-κB p65 Feedback Regulation Promotes Chronic Myeloid Leukemia Blast Crisis. Front. Oncol. 2021, 11, 654411.
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118.
- Borkiewicz, L.; Kalafut, J.; Dudziak, K.; Przybyszewska-Podstawka, A.; Telejko, I. Decoding LncRNAs. Cancers 2021, 13, 2643.
- Mishra, K.; Kanduri, C. Understanding Long Noncoding RNA and Chromatin Interactions: What We Know So Far. Non-Coding RNA 2019, 5, 54.
- Guh, C.-Y.; Hsieh, Y.-H.; Chu, H.-P. Functions and properties of nuclear lncRNAs-from systematically mapping the interactomes of lncRNAs. J. Biomed. Sci. 2020, 27, 44.
- Zhang, Y.; Liu, Q.; Liao, Q. Long noncoding RNA: A dazzling dancer in tumor immune microenvironment. J. Exp. Clin. Cancer Res. 2020, 39, 231.
- Saxena, A.; Carninci, P. Long non-coding RNA modifies chromatin. BioEssays 2011, 33, 830–839.
- Shi, T.; Guo, D.; Xu, H.; Su, G.; Chen, J.; Zhao, Z.; Shi, J.; Wedemeyer, M.; Attenello, F.; Zhang, L.; et al. HOTAIRM1, an enhancer lncRNA, promotes glioma proliferation by regulating long-range chromatin interactions within HOXA cluster genes. Mol. Biol. Rep. 2020, 47, 2723–2733.
- Kang, X.; Zhao, Y.; Van Arsdell, G.; Nelson, S.F.; Touma, M. Ppp1r1b-lncRNA inhibits PRC2 at myogenic regulatory genes to promote cardiac and skeletal muscle development in mouse and human. RNA 2020, 26, 481–491.
- Fan, S.; Tian, T.; Lv, X.; Lei, X.; Yang, Z.; Liu, M.; Liang, F.; Li, S.; Lin, X.; Lin, Z.; et al. lncRNA CISAL Inhibits BRCA1 Transcription by Forming a Tertiary Structure at Its Promoter. iScience 2020, 23, 100835.
- Song, W.; Gu, Y.; Lu, S.; Wu, H.; Cheng, Z.; Hu, J.; Qian, Y.; Zheng, Y.; Fan, H. LncRNA TRERNA1 facilitates hepatocellular carcinoma metastasis by dimethylating H3K9 in the CDH1 promoter region via the recruitment of the EHMT2/SNAI1 complex. Cell Prolif. 2019, 52, e12621.
- Gao, Z.; Chen, M.; Tian, X.; Chen, L.; Chen, L.; Zheng, X.; Wang, H.; Chen, J.; Zhao, A.; Yao, Q.; et al. A novel human lncRNA SANT1 cis-regulates the expression of SLC47A2 by altering SFPQ/E2F1/HDAC1 binding to the promoter region in renal cell carcinoma. RNA Biol. 2019, 16, 940–949.
- Hu, Z.; Mi, S.; Zhao, T.; Peng, C.; Peng, Y.; Chen, L.; Zhu, W.; Yao, Y.; Song, Q.; Li, X.; et al. BGL3 lncRNA mediates retention of the BRCA1/BARD1 complex at DNA damage sites. EMBO J. 2020, 39, e104133.
- He, R.-Z.; Luo, D.-X.; Mo, Y.-Y. Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis. 2019, 6, 6–15.
- Khan, M.R.; Wellinger, R.J.; Laurent, B. Exploring the Alternative Splicing of Long Noncoding RNAs. Trends Genet. 2021, 37, 695–698.
- Pisignano, G.; Ladomery, M. Epigenetic Regulation of Alternative Splicing: How LncRNAs Tailor the Message. Non-Coding RNA 2021, 7, 21.
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358.
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283.
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388.
- Videira, A.; Beckedorff, F.C.; Dasilva, L.F.; Verjovski-Almeida, S. PVT1 signals an androgen-dependent transcriptional repression program in prostate cancer cells and a set of the repressed genes predicts high-risk tumors. Cell Commun. Signal. 2021, 19, 5.
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914.
- Lin, C.; Yang, L. Long Noncoding RNA in Cancer: Wiring Signaling Circuitry. Trends Cell Biol. 2018, 28, 287–301.
- Zhang, T.; Liu, H.; Mao, R.; Yang, H.; Zhang, Y.; Zhang, Y.; Guo, P.; Zhan, D.; Xiang, B.; Liu, Y. The lncRNA RP11-142A22.4 promotes adipogenesis by sponging miR-587 to modulate Wnt5β expression. Cell Death Dis. 2020, 11, 475.
- Liu, A.; Qu, H.; Gong, W.; Xiang, J.; Yang, M.; Zhang, W. LncRNA AWPPH and miRNA-21 regulates cancer cell proliferation and chemosensitivity in triple-negative breast cancer by interacting with each other. J. Cell. Biochem. 2019, 120, 14860–14866.
- Wang, S.; Cheng, M.; Zheng, X.; Zheng, L.; Liu, H.; Lu, J.; Liu, Y.; Chen, W. Interactions Between lncRNA TUG1 and miR-9-5p Modulate the Resistance of Breast Cancer Cells to Doxorubicin by Regulating eIF5A. OncoTargets Ther. 2020, 13, 13159–13170.
- Gong, J.; Shao, D.; Xu, K.; Lu, Z.; Lu, Z.J.; Yang, Y.; Zhang, Q.C. RISE: A database of RNA interactome from sequencing experiments. Nucleic Acids Res. 2018, 46, D194–D201.
- Schönberger, B.; Schaal, C.; Schäfer, R.; Voß, B. RNA interactomics: Recent advances and remaining challenges. F1000Research 2018, 7, 1824.
- Pinkney, H.R.; Wright, B.M.; Diermeier, S.D. The lncRNA Toolkit: Databases and In Silico Tools for lncRNA Analysis. Non-Coding RNA 2020, 6, 49.
- Yotsukura, S.; Duverle, D.; Hancock, T.; Natsume-Kitatani, Y.; Mamitsuka, H. Computational recognition for long non-coding RNA (lncRNA): Software and databases. Briefings Bioinform. 2017, 18, 9–27.
- Li, J.; Zhao, Y.; Zhou, S.; Lang, L.; Zhou, Y. Inferring lncRNA Functional Similarity Based on Integrating Heterogeneous Network Data. Front. Bioeng. Biotechnol. 2020, 8, 27.
- Seifuddin, F.; Pirooznia, M. Bioinformatics Approaches for Functional Prediction of Long Noncoding RNAs. Methods Mol. Biol. 2021, 2254, 1–13.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.7K
Revisions:
2 times
(View History)
Update Date:
12 Nov 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




