| Food |
Major Allergens |
Molecular Mass (kDa) |
Types of Proteins |
The Structure of Proteins |
Allergy Symptoms |
Reference |
| Fish |
Pan h 1 |
10–13 |
Calbindin |
Contains 3 EF-hand regions (a motif composed of a 12-residue loop with a 12-residue-α-helix domain on each side), 2 of which can bind calcium. |
Blushing, hives, nausea, stomach pain, and intestinal bleeding. |
[31] |
| Shellfish |
Cra c 1 |
33–39 |
Protein bound to actin |
Adopting an α-helix structure, two molecules are entangled with each other to form a parallel dimeric α-helix structure. |
Nausea, diarrhea, abdominal pain, and muscle paralysis. |
[32] |
| Cra c 2 |
38–45 |
Phosphoglycoprotein |
Arginine kinase consists of an N-terminal domain (1–111) and a C-terminal domain (112–357). The N-terminal domain is all α-helices, and the C-terminal domain is an 8-strand anti-parallel β-sheet structure surrounded by 7 α-helices. |
[33] |
| Milk |
Bos d 8 |
57–37.5 |
Phosphate calcium binding protein |
Consists of 4 independent proteins: αs1-casein, αs2-casein, β-casein, and κ-casein. |
Skin rash, urticaria, eczema, vomiting, diarrhea, abdominal cramps, etc. |
[34] |
| Bos d 4 |
14.4 |
Combine with metal ions and participate in lactose synthesis |
With a two-piece structure containing α-single loop and 310 helix larger subdomain. |
[35] |
| Bos d 5 |
18 |
Lipid transporter |
Consists of two subunits connected by non-covalent bonds, mainly in the form of dimers. |
[36] |
| Egg |
Gal d1 |
28 |
Phosphoglycoprotein |
Contains 3 independent homologous structural energy domains, and 3 functional domains are arranged consecutively in space. |
Eczema, dermatitis, urticaria, vomiting, diarrhea, gastroesophageal reflux, etc. |
[37] |
| Gal d2 |
45 |
Phosphoglycoprotein |
Containing 4 free sulfhydryl groups, composed of 385 amino acid residues, these amino acid residues are twisted and folded to form a spherical structure with high secondary structure, most of which are α-helix and β-sheet. |
[38] |
| Gal d3 |
77 |
Iron-binding glycoprotein |
Consisting of 686 amino acids, including 12 disulfide bonds, the N-terminal and C-terminal 2 domains each contain a binding site for Fe3+. |
[39] |
| Gal d4 |
14.3 |
Basic globulin |
A single peptide chain composed of 18 kinds of 129 amino acid residues, with 4 pairs of disulfide bonds to maintain the enzyme configuration, with lysine at the N-terminus and leucine at the C-terminus. |
[40] |
| Peanut |
Ara h 1 |
63.5 |
7S Globulin |
The secondary structure contains β-turns, and the quaternary structure is a trimeric complex formed by 3 monomers. |
Angioedema, hypotension, asthma, anaphylactic shock, etc. |
[41] |
| Ara h 2 |
17–20 |
2S Albumin |
A monomeric protein. |
[42] |
| Ara h 3 |
57 |
11S Globulin |
The N-terminal and C-terminal domains of the monomer form contain 2 ciupin folds (composed of two sets of parallel β-turns, random coils and 3 α-helices). |
[43] |
| Wheat |
Tri a 36 |
40 |
Gluten |
- |
Wheat exercise stimulates allergies, urticaria, dermatitis, bread asthma, nausea, and diarrhea. |
[44] |
| Soybean |
Gly m 5 |
150–200 |
7S Globulin |
Trimer composed of α’-subunit, α-subunit and β subunit. |
Red and itchy skin, asthma and allergic rhinitis, abdominal pain, diarrhea, etc. |
[45] |
| Gly m 6 |
320–360 |
11S Globulin |
A hexamer composed of the interaction of G1, G2, G3, G4, and G5 subunits. |
[46] |
| Nuts |
Ana o 1 |
50 |
7S legumin |
Exist as a trimer in natural state. |
Metallic taste in the mouth, edema of the tongue or throat, difficulty breathing and swallowing, urticaria all over the body, flushing of the skin, cramping abdominal pain, nausea. |
[47] |
| Jug r 2 |
44 |
Consists of 593 amino acid residues. |
[48] |
| Cor a 11 |
48 |
Consists of 401 amino acid residues, with two potential N-glycosylation sites (Asn38 and Asn254) and a leader peptide of 46 amino acids. |
[49] |
| Ana o 3 |
14 |
2S albumin |
Composed of 5 helical structures, containing 2 subunits, connected by cysteine disulfide bonds. |
[50] |
| Jug r 1 |
15–16 |
Consists of 142 amino acid residues. |
[51] |
| Jug r 4 |
58.1 |
11S globulin |
Except for the first 23 amino acid residues which are predicted as signal peptides, the remaining part has a total of 507 amino acid residues. |
[52] |
| Cor a 9 |
40 |
Composed of 515 amino acid residues, the sequence homology with Ara h 3 is about 45%. |
[53] |
| Pru du 6 |
350 |
Exist in the form of hexamers, each monomer subunit is composed of one acid chain of 40 to 42 kDa and one alkaline chain of 20 kDa. |
[54] |
3. Detection of Animal Food Allergens
Seafood allergy is not only an important public health issue, but a serious food safety issue that affects the quality of life and may even be life threatening
[55]. For people with seafood allergies, avoiding foods containing seafood allergens is still the best option. Therefore, the monitoring of allergens is a process that requires strict supervision
[56]. In order to evaluate seafood allergens, new detection methods with high sensitivity and high efficiency are required.
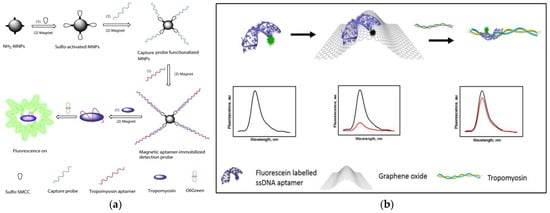
As we all know, magnetic separation is easy to operate and can effectively reduce or eliminate the interference from complex matrices in food. Therefore, based on functionalized magnetic nanoparticles (MNPs) as a separation carrier, Zhang et al. developed a simple and versatile label-free aptamer-based fluorescent sensor for the sensitive detection of TM (
Figure 1a)
[57]. In the study, OliGreen dye was selected as a fluorescent signal probe. The aptamer hybridizes with the capture probe bound to the surface of the MNPs to form an aptamer-MNPs complex as detection probe. When interacting with the target, the conformation of the complex changes, resulting in the release of the aptamer from the surface of the MNPs. So, the released aptamer in the supernatant produced a significant fluorescence enhancement signal, which is because the combination of OliGreen dye and ssDNA will produce ultrasensitive and specific fluorescence enhancement phenomenon. It is worth noting that when the commercially available OliGreen dye is in the free state, the fluorescence is weak or no fluorescence, but the fluorescence will increase by more than 1000 times once combined with the aptamer ssDNA. Under the optimal conditions, the linear range was 0.4–5 μg mL
−1 (R
2 = 0.996), with a limit of detection LOD of 77 ng mL
−1. In addition, the highly selective aptamer-based fluorescent sensor was successfully applied to the detection of TM in food matrix. Wu et al. also developed a similar sensor with a LOD of 4.2 nM and the concentration linear from 0.5–50 μg mL
−1 [58]. Recently, Chinappan et al. developed an aptamer-based fluorescent-labeled sensor for the detection of TM. (
Figure 1b)
[59]. Graphene oxide (GO) is used as a platform for screening the minimum length of aptamer sequences that can bind to the target with high affinity. A fluorescein dye labeled GO quenches the truncated aptamer by π-stacking and hydrophobic interactions. After the addition of TM, the fluorescence was restored due to the competitive binding of the aptamer to GO. More importantly, the aptamer selected in this study is a truncated ligand fragment, which has four times higher affinity than the full-sequence aptamer, with a LOD of 2.5 nM. The developed aptamer-based fluorescence sensor can complete the detection within 30 min. The performance of the sensor was confirmed in the addition experiment of chicken broth, and a high percentage recovery rate (~97 ± 10%) was achieved. Compared with the above studies, the sensitivity and specificity of this work have been greatly improved.
Figure 1. (
a) Schematic of preparation of magnetic-assisted fluorescent aptamer for tropomyosin detection. Reproduced with permission from
[57]. Copyright Sensors and Actuators B-Chemical, 2018. (
b) Schematic of graphene oxide-based fluorescent aptamer biosensor for TM detection A: Changes in the fluorescence intensity of the aptamer released from the GO surface; B: The linear correlation of the fluorescence intensity of TMT2 (at 515 nm) with the concentration of TM. Reproduced with permission from
[59]. Copyright Food Chemistry, 2020.
Fluorescence resonance energy transfer (FRET) is a mechanism widely used in the preparation of biosensors, which is an energy transfer phenomenon between two fluorescent molecules that are very close
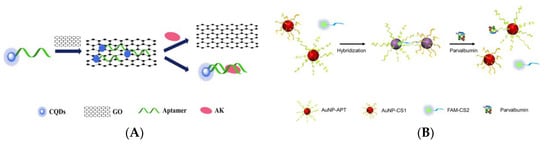
[60]. Zhou et al. designed an aptamer-based “on-off-on” fluorescent biosensor based on FRET and used developed carboxyl functionalized carbon quantum dots (cCQDs) and GO for the detection of shellfish allergen arginine kinase (AK) (
Figure 2A)
[61]. The cCQDs-aptamer probe and GO self-assemble for the first time through a specific π-π interaction, so that the fluorescence of cCQDs is effectively quenched. After the addition of AK, cCQDs-aptamer is released from the GO surface and then forms the cCQDs-aptamers-AK complex, which restores the fluorescence of cCQDs. The aptamer-based FRET sensor can perform sensitive detection in the AK concentration range of 0.001–10 μg mL
−1, with a LOD of 0.14 ng mL
−1 (S/N = 3) and a limit of quantification (LOQ) of 0.27 ng mL
−1 (S/N = 10). Furthermore, in a control experiment with a blank sample, it was found that the sensor has high specificity. This reliable, precise, highly specific, and easy-to-operate aptamer sensor may provide a new perspective for the application of fluorescence sensing technology in the field of food safety.
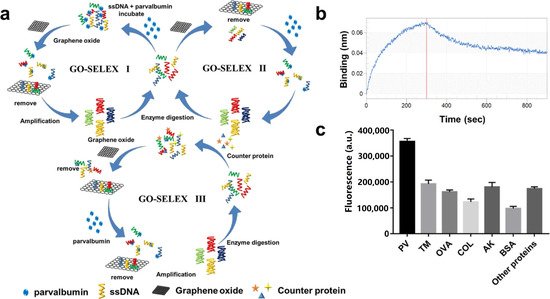
Figure 2. (
A) Schematic of a “on-off-on” fluorescence aptasensor for AK detection. Reproduced with permission from
[61]. Copyright Microchemical Journal, 2020. (
B) Schematic of a dual-mode fluorescence sensor for PV detection based on AuNP color changes and FAM-CS2 fluorescence changes. (
C) a: Schematic of the aptamer selection procedure by capturing GO-SELEX; b: Affinity of Apt5 towards PV; c: Specificity of Apt5 towards PV. Reproduced with permission from
[62]. Copyright Microchemical Journal, 2020.
In recent years, biosensors based on dual signals or functions have received widespread attention due to the diversity of detection. Dual-mode nanosensors usually use colorimetric and fluorescent reporters to achieve convenient visual inspection and highly sensitive fluorescent detection
[63][64]. Wang et al. developed a dual-mode aptamer-based fluorescent sensor for the detection of PV, the major allergen of fish (
Figure 2B)
[62]. In
Figure 2C(a), aptamer towards PV was obtained by in vitro screening of random ssDNA library containing a 40-mer randomized region using the triple-mode GO-SELEX. The aptamer-modified gold nanoparticle (AuNP-APT), complementary short-strand modified gold nanoparticles (AuNP-CS1), and fluorescent dye-labeled complementary short-strands (FAM-CS2) were assembled by DNA hybridization. After the addition of PV, the competitive interaction with aptamer leads to the decomposition of the aptamer sensor, resulting in the color shift of the AuNPs solution and the recovery of the FAM-CS2 fluorescence signal. The results showed that the aptamer sensor showed a good colorimetric response (2.5–20 μg mL
−1) and linear fluorescence correlation (2.38–40 μg mL
−1) in the PV concentration range. In addition, the affinity and specificity of the aptamer sensor were also investigated, as shown in
Figure 2C(b,c). Therefore, aptamer 5 with good affinity (KD = 7.66 × 10
−7 M) and specificity is the best aptamer for aptasensor construction. They also studied the feasibility of aptamer sensor in real fish samples, revealing the potential in field of monitoring and quantitative detection of food allergens.
Recently, as an alternative to antibodies, the use of peptide aptamers as biosensors has attracted more attention. Peptide aptamers usually contain 10–20 amino acids, which the high selective recognition ability is equivalent to that of antibodies. Phadke et al. used ribosome display technology to select two fluorescent peptide aptamers Cas1 and Cas2. for the detection of α-casein
[65]. Among them, 7-nitrobenzofurazan (NBD)-modified aminophenylalanine is coupled to the translated peptides to prepare fluorescent peptide aptamers. This is because the peptide can quench the fluorescence of NBD. Once the peptide recognizes the target α-casein, the NBD-modified phenylalanine is released, and its fluorescence will instantly increase. It is worth noting that although the fluorescence of the two aptamers increased slightly in the presence of the control protein β-lactoglobulin, the modification of Cas1 with polyethylene glycol (PEG-Cas1) inhibited this phenomenon, which is because PEG-Cas1 may inhibit the interaction between aptamer and β-lactoglobulin. The aptamer sensor with a LOD of 0.04 mM, is equivalent to that of the kit. Moreover, the system can detect α-casein in short time (20–25 s) when compared with the 15 min required by immunochromatography kits. In addition, it is found that when using PEG-Cas1 to detect casein, the instant increase in fluorescence can be observed even with the naked eye. This study has contributed to improving the specificity of aptamers.
In order to reduce the incidence of milk allergy, hypoallergenic formula (HF) has been commercialized as a substitute for milk
[66]. Nevertheless, in some cases, infants who consume these formula milk powder still have allergic reactions because of residual β-lactoglobulin in HF
[67]. Therefore, it is necessary to establish a method that can detect the lower concentration of β-lactoglobulin. Shi et al. used carbon dots (CDs) as a fluorescent signal and Fe
3O
4 NPs as a magnetic separator to establish a fluorescent-labeled assay for the detection of β-lactoglobulin
[68]. The assay is based on the hybridization between aptamers immobilized on Fe
3O
4 NPs and CDs-labeled complementary oligonucleotides (cDNA). In the presence of β-lactoglobulin, the aptamer preferentially binds to β-lactoglobulin, and part of the CDs-cDNA is released into the solution. After magnetic separation, the fluorescence signal of the supernatant increased with the increase of β-lactoglobulin concentration. Based on this, the aptamer assay with the range of 0.25–50 ng mL
−1 and a LOD of 37 pg mL
−1 has been successfully applied to the detection of trace β-lactoglobulin in HF. In the study of Qi et al., the binding mechanism of aptamer and β-lactoglobulin and the detection principle of fluorescent surface-enhanced Raman scattering (fluorescent-SERS) dual-mode aptamer sensor were thoroughly studied, which provides a theory basis and application potential for the development of aptamer sensors
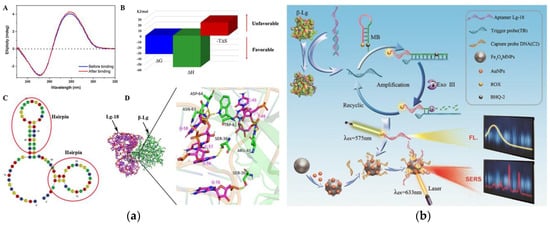
[69]. In
Figure 3a, the circular dichroism of Lg-18, thermodynamic parameters analysis, secondary structure of Lg-18, and the result of molecular docking between aptamer Lg-18 and β-lactoglobulin were performed to illustrate the successful selection of aptamer. The specific response principle of the dual-mode aptamer sensor is shown in
Figure 3b. The fluorescent-SERS aptamer sensor shows a wider linear range (10–5000 ng mL
−1), and the LOD is 0.05 ng mL
−1. Furthermore, under the interference of other proteins, the aptamer sensor showed excellent specificity.
Figure 3. (
a) Schematic of Lg-18 and β-lactoglobulin binding. A: Circular dichroism analysis of Lg-18 before and after binding. B: Analysis of thermodynamic parameters in the combined process. C: Secondary structure of Lg-18 predicted by Mfold online software. D: Analysis of molecular docking results of aptamer Lg-18 and β-lactoglobulin (
b) Schematic of aptamer-based fluorescent Raman dual-mode biosensor for detection of β-lactoglobulin. Reproduced with permission from
[69]. Copyright Sensors and Actuators B-Chemical, 2021.
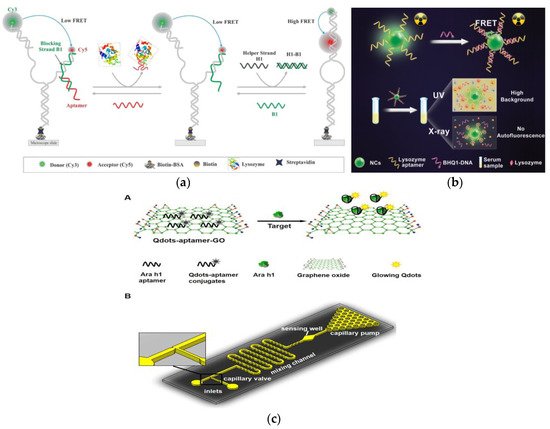
Lys, as an allergen in egg and a biomarker of many diseases, its detection and quantification are of great significance in clinical diagnosis
[70]. Sapkota et al. developed an aptamer sensor based on single-molecule FRET (smFRET) for the detection of Lys (
Figure 4a)
[71]. One of the arms has a blocking chain (B1), which is extended by 15 nucleotides to partially hybridize to the aptamer. The aptamer sensor remains open and almost no FRET efficiency occurs when Lys is not detected. After the addition of Lys, the aptamer binds to Lys and is displaced from the sensor, resulting in a foothold-mediated replacement of B1 by another chain, H1. At this time, the binding of Lys triggers the conformational transition state from low FRET to high FRET. Using this strategy, they demonstrated that the aptamer sensor can detect Lys at concentration as low as 30 nM, with a dynamic range extends to ~2 μM, and is almost free of interference from similar biomolecules. In addition, the smFRET method requires only a small number of aptamers, which offers the advantage of cost-effectiveness. In fluorescence detection, the presence of background fluorescence induced by ultraviolet-visible light in biological samples can lead to inaccurate detection results
[72]. However, Ou et al. developed an X-ray nanocrystal scintillator aptamer sensor for sensitive detection of Lys based on the characteristics of weak scattering and almost no absorption of biological chromophores under X-ray irradiation (
Figure 4b)
[73]. In this study, aptamer-labeled lanthanide-doped nanocrystalline scintillators are designed to detect Lys quickly and sensitively through FRET. The use of low-dose X-rays as the excitation source and nanocrystals containing heavy atoms can achieve efficient luminescence, which endows the aptamer fluorescence sensor with high sensitivity (LOD: 0.94 nM), specificity, and sample recovery. In addition, this technology can provide a new generation of high-efficiency strategy without autofluorescence interference for the sensing and detection of biomarkers in biomedical applications.
Figure 4. (
a) Schematic of aptamer-based fluorescent biosensor for Lys detection. Reproduced with permission from
[71]. Copyright sensors, 2020. (
b) Schematic of aptamer sensor based on nanocrystal scintillator for detecting Lys without autofluorescence. Reproduced with permission from
[73]. Copyright Analytical Chemistry, 2019. (
c) A: Schematic of the Qdots-aptamer-GO quenching sensing principle; B: Schematic of the designed microfluidic chip. Reproduced with permission from
[74]. Copyright Biosensors & Bioelectronics, 2016.