1. Beer, a Fermented Alcoholic Beverage
Globally considered as one of the oldest and most important alcoholic beverages, beer corresponds to the third most popular drink among consumers, after coffee and tea
[1][2][3]. According to the World Health Organization (WHO), beer dominates the second position on the scale of the most consumed alcoholic beverages worldwide, representing 34.3% of the total alcohol consumed, being preceded solely by spirits (44.8%)
[4]. The designation of beer derives from the Latin term “
biber”, which means “drink”, and it is defined as a fermented beverage obtained from starch and flavored with hops
[5][6][7].
Up to date, there are four base natural products needed to produce beer, namely malt, yeasts, hops and water
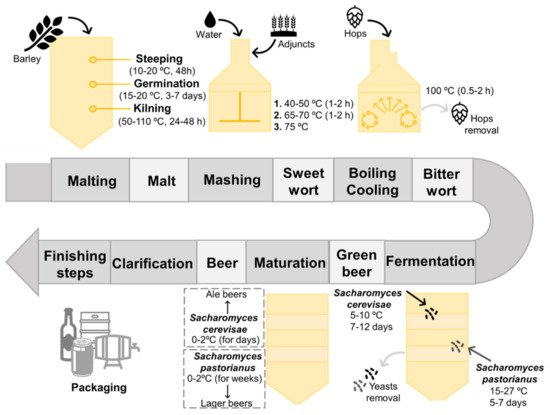
[5][7]. The set of processes involved in the production of beer is termed brewing, being summarized in
Figure 1 [3][7]. Malting corresponds to the first phase of brewing, comprising a set of stages, including steeping, germination and kilning, which lead to the production of malt from raw cereal grains
[2][8][9]. Barley (Hordeum vulgare) is the most frequently used cereal in beer production, since it can grow easily, even in adverse climate conditions; can be easily converted into malt; displays a suitable protein concentration (10–12%) required to yeast growth and foam production; and exhibits a significant carbohydrate content of 78–83%, of which 63–65% correspond to starch (one of the main sources of fermentable sugars used by yeasts during their fermentative metabolism)
[6][7][10]. Barley grains are initially steeped in aerated water conditions to increase their humidity, and then they are germinated at temperatures fluctuating from 15 to 20 °C, for 3 to 7 days
[2][10]. During this stage, several biochemical modifications occur in the grains, resulting on the synthesis of a variety of enzymes that are responsible for the breakdown of endosperm structural components, starch and proteins
[6][11]. After germination and subsequent removal of the sprouts, a kilning stage takes place to yield dry malted grains, in which controlled high temperatures are employed to reduce the water content of the grains without inactivating the enzymes, along with the occurrence of several Maillard reactions that are responsible for the generation of color and flavor-related compounds
[2][6][10][11][12]. In sum, the resultant cereal malt consists in a mixture of different components with a crucial importance in the next steps of brewing, such as saccharides, proteins, free amino nitrogen (FAN), enzymes and purine compounds, being considered the major contributors to the sensorial characteristics of the finished beer
[10][12].
Figure 1. Schematic overview of the brewing process.
During the second phase of brewing, termed mashing, the previous obtained malt is mixed with boiling water, followed by a resting period under regulated temperatures to enhance the activity of enzymes
[3][5][6][10][11]. Despite the popularity of barley, other substrates, acting as malt adjuncts, can be also introduced during this stage, for instance rice, sorghum, corn, wheat, sugars, syrups, etc., contributing not only to supply starch and sugar, but also to the development of specific attributes and leading to a wide diversity of beers
[6][10]. The temperature is initially maintained between 40 and 50 °C to enhance α-amylases, followed by an increment to temperatures between 65 and 70 °C to stimulate ß-amylases
[10]. At this time, we succeed in the hydrolyzation of ß-glucans by ß-glucanases, the hydrolyzation of proteins into their amino acids and peptides through the action of proteases, and the hydrolyzation of starch by α-amylases and ß-amylases into fermentable sugars, with maltose being the most abundant
[5][10][11][13]. Afterwards, the temperature is increased to at least 75 °C to inactivate the former enzymes, followed by a filtration step, resulting in a sweet wort that is defined as a complex mixture of fermentable sugars and other nutrients necessary for yeasts metabolism
[5][7][9][10]. The resultant wort is then transferred to a heating container (also known as brewing kettle) and hops are added, followed by a boiling stage for 1 to 2 h
[3][5][10][11][14]. Hops corresponds to the flower of the
Humulus lupulus plant, contributing to the aroma, flavor and stability of the finished beer due to the presence of several essential oils and resins (e.g., α-acids or humulone, ß-acids or lupulone, and tannins), some of them responsible for the aroma and bitterness profile of beer
[10][11][14][15]. During the boiling step, several events take place: (1) microorganisms and enzymes that remained active at the end of the mashing phase are killed and inactivated, respectively; (2) certain undesirable compounds are precipitated and removed; (3) resins and essential oils are extracted from hops, allowing the conversion of α-acids into their soluble form to improve the bitterness character, concentrate the flavor, and sterilize the wort; (4) color-related attributes are developed; (5) the remaining water is evaporated; and (6) the resultant wort is concentrated
[10][11][14]. At the end of this phase, wort is separated from the flavor agents, followed by a cooling step, resulting in a bitter wort that is transferred to a fermentation vessel
[5][10]. The next step in brewing is focused on the wort inoculation with yeasts, initially under aeration conditions to promote their growth, followed by an alcoholic fermentation process in which ethanol and carbon dioxide are formed
[16][3][5][10][11]. As stressed before, other by-products can be obtained from the yeast metabolism; however, they are obtained at lower concentrations
[16][17].
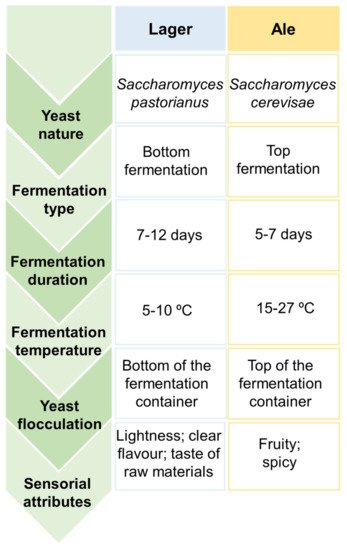
Depending on the nature of yeasts that remained at the end of the later process and the temperatures applied, two types of fermentation can be distinguished, particularly top and bottom-fermentations, that result on the rise of two main classes of beer, known as Ale and Lager, with the latter being the most common
[16][2][3][5][15]. The main differences between these two beer styles are summarized in
Figure 2.
Figure 2. Main differences between Lager and Ale beers.
Top-fermentations are characterized by being a rapid process performed by top-fermenting yeasts, such as strains of
Saccharomyces cerevisiae, under high temperatures ranging from 18 to 27 °C for 5–7 days
[5][14][15]. This kind of yeast has a predisposition to rise to the surface of the wort, adhere to carbon dioxide molecules, and subsequently produce foam. In contrast, bottom-fermentations are known by being a slow process accomplished by bottom-fermenting yeasts, such as strains of
Saccharomyces carlsbergensis (also known as
Saccharomyces pastorianus) and
Saccharomyces uvarum, that act under low temperatures ranging from 6–15 °C for 7–12 days
[10][14]. At the end of the fermentation process, these latter yeasts tend to flocculate and sink to the bottom of the wort. Therefore, top-fermentation are involved in Ale beers production, which is characterized by fruity and spicy attributes given by specific aroma-related compounds (e.g., esters, volatile phenols, etc.), while bottom-fermentation is associated to Lager beers manufacture, which is characterized by a lightness and cleaner flavor
[2][5][14].
After fermentation takes place, yeasts are removed, resulting in a non-consumable beer, known as green beer
[5]. Therefore, a maturation step must occur, under temperatures ranging from 0 to 2 °C for several days; however, the time required for maturation depends on the intended beer (e.g., lager beers take longer times to be aged than ale beers)
[5][10]. This stage allows not only the removal of undesirable compounds but also the development of the final flavor, color, and other attributes. Certain brewery industries include also a conditioning step, under temperatures of −1 °C, for at least 3 days, allowing the clarification through the removal of the remaining yeast, proteins, and other suspended solids
[10][14]. The ultimate phase of brewing is focused on the proper preparation of beer to be commercialized. Depending on the desired final product, distinct processing steps, some of them optional, are applied: (1) addition of proteases to counteract haze generation by residual proteins, during storage under refrigeration temperatures; (2) clarification and filtration to remove the remaining suspended solids; and (3) adjustment of carbon dioxide level (0.45–0.52%) by a carbonation method that implies the addition of this molecule to the final product or, as an alternative, the addition of a fresh yeasted wort to allow the occurrence of a secondary fermentation
[10]. The finished beer can be packaged into cans, bottles, kegs, and/or barrels. However, before being placed on the market, certain beers, particularly those that require a longer conservation period, such as canned and bottled beers, are subjected to an additional treatment, usually pasteurization, to improve their shelf-life, while others require a higher period of inoculation with yeasts to enhance a secondary fermentation process and, consequently, to endorse the production of by-products responsible for specific flavors
[5][10][11].
The chemical analysis of beer is crucial to determine and guarantee its quality and safety, but also to understand its nutritional aspects and organoleptic attributes
[7]. Apart from water, which is the major constituent representing around 90% of the final product, beer presents other molecules, namely carbohydrates, ethanol, and carbon dioxide
[6][18]. Despite most of the sugar present in wort being fermented into ethanol during brewing, some non-fermentable carbohydrates remain at the end of the process
[6]. Carbohydrates represent 3.3 to 4.4% of the final product, of which 75–80% correspond to dextrins, 20–30% to mono and oligosaccharides, and 5–8% to pentosans
[18]. Ethanol is considered the most important alcohol in beer that also acts as a flavor and sweetness enhancer, being found at concentrations that vary according to the type of beverage
[6][7]. For instance, it can range from 20,000 to 80,000 mg.L
−1, with a percentage of alcohol per volume variable from 0.05% in alcohol-free beers to 12.5% in British Thomas Hardly Ale (the strongest beer worldwide)
[7]. Carbon dioxide is typically found at concentrations ranging from 3.5 to 4.5 g·L
−1; however, certain types of beers, such as those that are bottle conditioned, can exhibit higher contents
[6]. Carbon dioxide is a result of yeast metabolism, which impacts foam generation and the flavor profile of the finished product since it assures the proper delivery of volatile compounds into the headspace of beers. Along with these major compounds, beer encompasses other circa 800 organic and non-organic compounds, being present in smaller amounts. Some of them are derived from raw materials and do not suffer any alteration during brewing, while others are produced as by-products due to yeasts metabolism. The main constituents of beer, their contents, and sources are summarized in
Table 1.
Table 1. Chemical composition of regular beer (information provided in the literature
[6][7][18].
| |
Concentration |
Source |
| Water |
90% |
- |
| Ethanol |
20,000–80,000 mg L−1 |
Yeast, malt |
| Carbon Dioxide |
3500–4500 mg L−1 |
Yeast, malt |
| Carbohydrates |
3.3–4.4% |
Malt |
| Inorganic salts |
500–2000 mg L−1 |
Water, malt |
| Total nitrogen compounds |
300–1000 mg L−1 |
Yeast, malt |
| Organic acids |
200–500 mg L−1 |
Yeast, malt |
| Higher alcohols |
60–100 mg L−1 |
Yeast, malt |
| Aldehydes |
10–20 mg L−1 |
Yeast, hops |
| Esters |
60–80 mg L−1 |
Yeast, malt, hops |
| Sulphur compounds |
1–10 mg L−1 |
Yeast, malt, hops |
| Hop derivatives |
20–60 mg L−1 |
Hops |
| Complex B vitamins |
5–10 mg L−1 |
Yeast, malt |
Inorganic salts, derived from malt and/or water used during brewing, can be found at concentrations ranging from 500 to 2000 mg.L
−1, comprising cations, such as magnesium and sodium; anions, such as chloride and sulfate; and trace metals
[6][7][18]. Depending on their nature and concentration, these elements can produce negative or positive effects on the sensorial profile of the finished beer, influencing the taste and clarity properties
[6][7]. Several alcohols can also be present, such as higher alcohols (e.g., 3-methylbutanol, 2-methylbutanol, 2-methyl-propanol, propanol and ß-phenylethanol) at concentrations ranging from 60 to 100 mg.L
−1; contents higher than 100 mg.L
−1 can led to negative effects on flavor and need to be regulated
[6]. Some phenolic alcohols can also be found, being derived from the breakdown of malt and from hops polyphenols. These are responsible for the texture and harshness, as well as glycerol derived from yeast metabolism, which play an important role on taste, enhancing smoothness and mouthfeel. Organic acids (e.g., acetic acid, lactic acid and succinic acid), which are derived from yeast metabolism through deamination reactions of the amino acids present in wort, can also be found at concentrations ranging from 200 to 500 mg.L
−1 [6][18]. These compounds are responsible for the relative acidity of beer (normal pH of 4.0–5.0; however, some beers have a pH around 3.0). Small amounts of vitamins can be found, namely complex B vitamins (e.g., biotin, inositol, pantothenic acid, etc.), that act as growth factors assimilable by yeast to ensure their performance, justifying their low contents on beer
[6][7]. Esters and aldehydes, generated through esterification and dehydrogenation reactions of alcohols, respectively, are also present
[6]. Esters (e.g., ethyl esters) can be found at concentrations ranging from 60 to 80 mg.L
−1 and are related to the fruity and floral aromas, while aldehydes (e.g., acetaldehyde) are present with contents ranging from 10 to 20 mg.L
−1, being associated with the characteristic young flavor of green beer. Beer additionally presents hop derivatives, such as iso-α-acids, at concentrations ranging from 10 to 100 mg·mL
−1 [7]. They are considered the primary flavor constituents, after ethanol and carbon dioxide, being responsible for the beer bitterness, foam stabilization and inhibition of microbial growth. Sulphur compounds (e.g., sulfur dioxide, hydrogen sulfite, etc.) derived from malt, hop and yeast metabolism, are present at concentrations ranging from 1 to 10 mg.L
−1, being responsible for the overall flavor profile of beer, while allowing the monitorization of the processing operations involved in brewing
[6][7]. Finally, beer possesses a wide range of nitrogenous compounds, found with a content ranging from 300 to 1000 mg.L
−1, of which 80–85% are derived from malt and the remaining from yeast metabolism
[6][19]. This comprises amino acids, peptides, polypeptides, amines, nucleic acid and derivatives, heterocyclic compounds, and particularly metabolites that result from nucleic acids catabolism, such as purine and pyrimidine compounds. During the mashing phase, proteins and peptides are catabolized into their amino acids, resulting in high contents of nitrogenous compounds available to the yeast growth. Therefore, most of the amino acids present in the finished beer correspond to those that were not assimilated or used by yeast to produce higher alcohols
[6]. Moreover, during malting and mashing phases of brewing, nucleic acids are enzymatically catabolized into their respective purine and pyrimidine nucleotides, nucleosides and free bases, contributing to their appearance in the finished beer
[6][7][19]. Little is known regarding pyrimidine compounds in beer; however, the presence of purine compounds in this beverage is well known and documented
[6].
2. Removal of Purine Compounds from Beer
Beer consumption represents a risk factor of gout, mainly due to its purine content; therefore, a reduction on the intake of these molecules is considered an effective practice to prevent and manage the disease
[20][21][22][23]. In this sense, it is of extreme importance to develop techniques that allow the removal of purine compounds from food products. This possibility will assist hyperuricemic and gouty patients to achieve a proper management of the disease, avoiding the common restrictive dietary methods, while enabling them to maintain serum uric acid concentrations within the reference value and reduce clinical manifestations. Accordingly, efforts have been carried out to reduce the purine content in food items, being focused on enzymatic degradation or on adsorption methods, using a variety of adsorbent materials, which are overviewed below.
3.1. Enzymatic, Biological and Processing Methods
The application of enzymes or microorganisms in food products is an old process, experiencing a significant increase in recent years
[24][25]. Despite their relevance, few methods were, however, reported by envisaging the reduction of the total purines content of malt-derived fermented beverages and foods
[26][27][28]. Given the limited number of works in this field, those applied to food items are also reviewed, and they could be of value to design strategies to remove purine compounds from beer.
Shibano et al.
[27] disclosed a brewing process to obtain beer with a reduced purine content. A nucleoside phosphorylase isolated from calf spleen was activated and applied to wort to allow the catabolism of purine nucleosides into their bases, which were further assimilated by yeast during fermentation. In wort without enzymatic treatment, it was verified a decrease on the amount of purine bases (e.g., adenine, guanine and xanthine), from 70 to 10 mg.L
−1, during the initial stage of fermentation, whereas no changes were detected on the amount of purine nucleosides (e.g., adenosine, guanosine and inosine; approximately 90 mg.L
−1). This proves that purine bases are uptake and assimilated by yeast during fermentation. However, after treating wort with the isolated enzyme, it was found that a decrease on the purine nucleosides contents, whereas an increase in the concentrations of bases was detected. A nucleoside decomposition ratio of around 60% was determined, proving the enzyme effectiveness on the decomposition of nucleosides. Since it was proved that purine bases are further assimilated by yeasts, the same compounds resultant from purine nucleosides decomposition could be also uptake and used, leading to a beer with a low purine content. This method is, however, dependent on the degradation rate of purine nucleosides and on the assimilation rate of the total purine bases and nucleosides initially present in wort
[29].
Mahor et al.
[30] demonstrated the possible role of a purine nucleoside phosphorylase (PNP) from
Kluyveromyces lactis (KlacPNP), an enzyme involved in the purine catabolism, and its variant KlacPNPN256E, in catalyzing a crucial reaction that leads to a reduction on the purine contents in beer. According to the results, after treating beer with these enzymes, a significant conversion of inosine into hypoxanthine by KlacPNP, as well as inosine to hypoxanthine and adenosine into adenine by KlacPNPN256E, occurred. These findings suggest the applicability of KlacPNP and respective variants on the crucial steps that contribute to decrease the purine contents in beer. Moreover, Mahor et al.
[31] used adenine deaminase and guanine deaminase, involved in the conversion of adenine to hypoxanthine and guanine to xanthine, respectively, of
Kluyveromyces lactis, to reduce the purines content in beer. A reduction on adenine contents of 66–67% and a decrease of guanine concentration from 68.8 µmol.L
−1 to residual values were found. Furthermore, when adenine and guanine deaminases were combined with other important degrading enzymes and applied in beer, a reduction of the total purine content was verified.
Jankowska et al.
[26] reviewed the role of a novel enzymatic approach, using
Arxula adeninivorans endogenous and recombinant purine degradative enzymes, in the manufacture of food products with low purine compounds content. This enzymatic system is composed of four purine-degrading enzymes, namely adenine deaminase, guanine deaminase, xanthine oxidoreductase and urate oxidase, that simultaneously catabolize purines into the water-soluble compound 5-hydroxyisourate and preventing uric acid accumulation. This enzymatic mixture was applied in a rolled fillet of ham, and it was verified that, after inoculation, adenine, hypoxanthine, xanthine, guanine, and uric acid contents were reduced. The authors
[32] used a purified recombinant
Arxula adeninivorans, capable to produce high amounts of adenine deaminase to achieve a beef broth with lower purine contents. This enzymatic methodology was able to reduce adenine contents from 70.4 to 0.4 mg.L
−1, paving the way for its use to reduce the production of uric acid. Moreover, Jankowska et al.
[33] used
A. adeninivorans, which is able to produce xanthine oxidoreductase to obtain food products with a reduced xanthine and hypoxanthine contents. It was found that there was a reduction from 169.24 to 39.59 mg.L
−1 in xanthine concentrations, along with a decrease from 171.56 to 52.22 mg.L
−1 in hypoxanthine contents. Although this mixture of enzymes was only applied to reduce the purine contents in food items, its use in fermented malt beverages, such as beer, could also be seen as a promising possibility.
Chen et al.
[28] proposed a method to reduce purine content of an edible material using specific microorganisms capable of digesting these molecules, followed by their removal through conventional separations methods (e.g., centrifugation and filtration). The microorganisms were selected from
Aspergillus Oryzae ATCC 11 493,
Aspergillus Oryzae ATCC 26831,
Aspergillus Oryzae ATCC 44193,
Rhizopus oryzae ATCC 52362 and combinations of both. This biological method was applied in water extracts of mushrooms and soy-bean milk, in which adenine, guanine, hypoxanthine and xanthine contents were reduced after treatment. Moreover, by using
Aspergillus Oryzae ATCC 26831 (1.3 × 106 spore/mL), a reduction of 84, 61 and 97% of the total purine content in three distinct mushroom water extracts, respectively, was verified after 48 h, whereas, by using the same microorganism and
Aspergillus Oryzae ATCC 44193 (1.3 × 106 spore/mL), a reduction of 15.24 and 39.93% in soy-bean milk, respectively, was verified after 16 h. These findings suggest the effectiveness of these microorganisms in reducing the total purine content of edible materials. This biological method could be also applied to reduce purine content of alcoholic beverages, such as beer, and as suggested by the authors.
Overall, and although scarcely studied, enzymatic and biological methods allow for the reduction of purine compounds’ content in foods and beverages, including beer. Nevertheless, the applicability of enzymatic methodologies in the food industry brings out additional challenges, such as the difficulty of enzymes to preserve their activity under conditions compatible with the food-storage conditions for long periods, and for which further research in this field is highly recommended.
3.2. Adsorption Methods
Given the limitations observed for enzymatic and biological methods, it is imperative to develop other methodologies that could allow an efficient removal of purine compounds from foods and beverages. This will allow the manufacture of products with low purine contents, enabling their consumption by hyperuricemic and gouty patients and the decrease of the risk to develop these diseases. Adsorption methods can be an efficient alternative towards these goals. Despite their potential, there are still few reports focused on the removal of purine compounds from matrices such as beer and wort, and these are dominated by adsorption processes that use carbon-based materials, namely activated carbon or charcoal
[29][34][35]. These works, as well as the previously mentioned enzymatic degradation methodologies for purines removal, are summarized in
Table 2.
Table 2. Removal-based methodologies of purine compounds from beer
[31][27][30][34][35].
| Target |
Removal Agent |
Method |
Matrix |
Application |
| Adenine, guanine, xanthine, adenosine, guanosine and inosine |
Nucleoside phosphorylase isolated from calf spleen |
Enzymatic
degradation |
Wort |
Brewing process for the manufacture of a beer reduced in purines |
| Inosine and hypoxanthine |
Purine nucleoside phosphosphorylase from K. lactis (KlacPNP) and KlacPNP256E variant |
Enzymatic
degradation |
Beer |
Reduction of the purine content of beer |
| Adenine and guanine |
Recombinant adenine and guanine deaminases of K. lactis |
Enzymatic
degradation |
Beer |
Reduction of the purine content of beer |
| Nucleic acid derivatives (e.g., guanine and guanosine) |
Activated carbon |
Adsorption |
Wort and beer |
Recover and concentration of nucleic acids |
| Adenosine, guanosine and guanine |
Activated charcoal prepared with beer lees |
Adsorption |
Beer and low-malt beer |
Production of a malt fermented beverage with reduced purine content |
Activated carbon or charcoal is a common adsorbent material due to its several advantages, i.e., high surface area, high adsorption capacity, rapid adsorption kinetics and relative easiness in being regenerated
[36]. For instance, Buday et al.
[34] reported the use of activated carbon to recover concentrated nucleic acid derivatives, such as guanine or guanosine, from biological samples, for instance, wort and beer. Fujino et al.
[29] proposed a method to obtain malt fermented beverage with a reduced purine concentration, in which these molecules were efficiently and selectively removed from wort or fermentation solution by using activated charcoal as an adsorbent material. It was found that an activated carbon with an average pore diameter of 1–3.5 nm and occurring as a fine powder is more effective in removing purine compounds (adenosine, guanosine, and guanine) from aqueous solutions. However, when applied to beer and low-malt beer samples, it was found that an activated charcoal with an average pore diameter of 1.8 nm, added in 500 mg/100 mL, is more effective, with an adsorption capacity for purine compounds ≥90%. The selectivity for the material for these molecules was also evaluated, with no alterations found in the contents of other compounds, such as ethanol and amino acids, proving the high selectivity and efficiency of the material to remove purines from beer and low-malt beer. Nevertheless, a decolorization event could occur in the resultant fermented malt beverages after treatment with activated charcoal.
Shibata et al.
[35] used activated carbon, prepared from brewing waste material, i.e., beer lees, to remove purine compounds from beer. A variety of beer lees-based activated carbons were prepared (e.g., LPN37, C950H1(2), C940H1.5(3) and C960H1.5(3)), and their adsorption capacity towards adenine, adenosine and AMP was evaluated and compared to those of a commercial activated carbon. It was found that the prepared beer lees-based activated carbon materials can remove the target molecules with an almost equal removal efficiency to that of the commercial activated carbon. It was also concluded that the total purine bases’ contents decrease with the increase in the amount of activated carbon employed. When 3 g/100 L beer of C940H1.5(3) or C960H1.5(3) was used, an almost 100% removal of the total amount of purine bases was accomplished; however, when the same amount of LPN37 or C950H1(2) was applied, this content decreases to 10–20 mg/L. These results shown that C940H1.5(3) and C960H1.5(3) present a higher adsorption capacity for purine compounds in beer. Nevertheless, there are some disadvantages associated with the use of this kind of materials. Due to their molecular sieve behavior, the adsorption ability decreases for relatively small purines (e.g., adenine), while for relatively large purines (e.g., AMP) the adsorption ability increases. Furthermore, these materials displayed low selectivity towards purines, since other molecules, such as amino acids, flavor-related compounds and essential nutrients, were simultaneously removed.
Given the lack of work focused on the removal of purine compounds from beer by adsorption methods, studies involving the adsorption of purines and related molecules from other matrices are also here overviewed, paving the way for its applicability in beer. To get solutions to better manage gout disease, several researchers developed various adsorbent materials able to remove purines and related compounds. Among them, microporous polymer granule
[37], zinc oxide nanoparticles loaded on activated carbon
[36] and polyethyleneimine/SiO
2 [38] were developed, and their potential to remove uric acid from blood samples, preventing the excessive serum uric acid content, was tested. For instance, the adsorption of uric acid onto zinc oxide nanoparticles loaded on activated carbon (ZnO-NP-AC) was studied, and the efficacy of this adsorbent was proven
[36]. A high adsorption capacity of 345.8 mg/g for the removal of uric acid was achieved. This finding suggests the applicability of activated carbon materials to adsorb and remove uric acid, which may prevent an excess of this metabolite. Additionally, Liu et al.
[39] studied the impact of a pitch-based spherical activated carbon, modified by chemical vapor deposition of NH
3, on uric acid adsorption properties. It was found that its adsorption occurs as a spontaneous, endothermic and irreversible process, thus suggesting its possible application to remove uric acid from blood samples. Since these adsorbent materials were able to efficiently remove uric acid from blood, resolving gout disease in a more evolved stage, i.e., after uric acid formation during purine catabolism, their applicability to remove purine molecules from beer could also be envisioned. It was also reported the use of dextrans
[40] and phyllosilicates
[41] as adsorbent materials to remove purines compounds and nucleic acids from aqueous solutions, respectively. Since beer is an aqueous solution presenting a panoply of components, including purine compounds, the applicability of these adsorbent materials to remove these target molecules from such matrices could also be useful and envisioned for this propose.
Given the features exhibited by activated charcoal materials, Tsurushima et al.
[42] proposed an adsorption method based on activated carbon to efficiently separate nucleotides and nucleosides from a solution containing both compounds. After adsorption, nucleotides were successfully eluted with an aqueous solution of an alkali metal hydroxide, proving the ability of activated carbon to adsorb these target compounds. Icenhour et al.
[43] proposed an adsorption procedure, using activated charcoal coated with polyvinylpyrrolidone, dextran or coconut flours, to extract nucleic acids, namely DNA and RNA, from complex matrices, such as stool and water samples.
Beyond activated carbons, other novel materials have been proposed for the adsorption of purine compounds and relatives, such as carbon nanotubes (CNTs) and graphene-based materials
[44][45]. CNTs, discovered in the 1960s, correspond to hollow cylindrical sheets of hexagonal carbon atoms networks, being considered a metallic or a semi conductive material
[44]. They exhibit a high surface area, accompanied by a small diameter and high curvature, allowing the establishment of effective interactions with target compounds, mainly through van der Waals forces, π–π stacking and hydrophobic interactions. Due to their advantages, some studies have been reported that proves the adsorption of nucleic acids and purine compounds on CNTs surface, although the majority of them focused on the evaluation of the interactions involved on the adsorption process
[46][47]. For instance, Yaroslav et al.
[47] studied the adsorption of adenine, thymine and their radicals on the surface of metallic and semiconducting single-wall carbon nanotubes. It was found that these molecules are physisorbed, mainly due to the interactions of their π-orbitals with the π-orbitals of the adsorbent. Moreover, Nandy et al.
[46] studied the adsorption of nucleic acids, namely single- and double-stranded DNA (ssDNA and dsDNA, respectively) on CNTs, demonstrating that single-stranded DNA (ssDNA) strongly adsorbs and wraps around CNT surface, with almost all nucleobases interacting with the material through van der Waals forces. The same group of researchers
[46] evaluated the adsorption of the same nucleic acid (ssDNA and dsDNA) on the surface of a graphene material.
Graphene emerged in 2004 and, since then, aroused great interest within the scientific community
[45][48]. In its pristine form, graphene corresponds to a two-dimensional (2D) material, formed by a thin layer of carbon atoms arranged in an sp
2-bonded aromatic structure with a one-atom thickness, also known as honeycomb crystal structure
[44][49]. Graphene presents unique physical, chemical and thermal properties, being considered an ideal material for a diversity of applications
[44][50]. Within its panoply of attributes, graphene exhibits a high surface area, which is related to its high capacity to adsorb organic molecules
[50][51][52][53]. Based on these features, Nandy et al.
[46] showed that dsDNA can be adsorbed on the surface of graphene, which is mainly due to non-covalent interactions, particularly π–π stacking. It was found that the binding affinity of the target molecule to graphene is higher, comparing to those obtained in CNTs. This can be explained by the advantageous properties exhibited by graphene materials, such as their high surface area and their easy surface modification
[49]. Moreover, Antony et al.
[54] studied the process and interactions energies involved in the adsorption of DNA nucleobases, including guanine (G), adenine (A), cytosine (C) and Thymine (T) on graphene, by dispersion-corrected density functional theory (DFT-D). It was found a binding crescent sequence as follows: G > A > T > C. Similarly, Sowerby et al.
[55] used a single solute adsorption isotherm method to study the adsorption process of the same DNA nucleobases onto the graphite-water interface, and it was found a similar adsorption strength crescent sequence (G > A > T > C). On the other hand, Gowtham et al.
[56] evaluated, through a plane-wave pseudopotential approach within the local density approximation (LDA) of DFT, the interaction energies involved in the adsorption of the same DNA nucleobases onto graphene, and they found an energy-binding sequence following the order G > A~T~C. Finally, Varghese et al.
[57] studied the adsorption process of the same DNA nucleobases onto graphene, and it was found that the binding energies involved are generally weak, exhibiting the sequence G > A > C~T. These findings prove that DNA nucleobases can adsorb onto graphene materials, the binding energies involved are generally weak, and guanine can establish a stronger interaction with the material when compared to the other nucleobases.
Nandy et al.
[46] studied the adsorption and interactions involved of single and double-stranded DNA (ssDNA and dsDNA, respectively) on a variety of materials, including CNTs, dendrimers and graphene, and it was found that dsDNA macromolecules adsorb better onto graphene. Liu et al.
[58] studied the adsorption of DNA onto gold nanoparticles and graphene oxide (GO), being found that the GO heterogeneous surface (due to the presence of hydrophobic regions) is favorable for DNA adsorption. Moreover, Lee et al.
[59] demonstrated the adsorption of DNA onto graphene, and it was found that besides π–π stacking, other interactions can take place, leading to an interfacial dipole interaction between nucleobases and graphene. These findings prove not only that DNA and its derivatives have the capacity of being adsorbed onto graphene materials, but also that this process is mainly driven by π–π stacking interactions.
The works regarding adsorption methods of purine and close compounds using a diversity of adsorbent materials are compiled in Table 3. It is important to reinforce that, although a majority of the exposed adsorption works focus on the study of the process and interactions involved, the high capability of these materials to adsorb purine molecules and close compounds was shown. These findings are highly important in the sense that they could pave the way for the application of these materials in the food industry to remove purine compounds from food items, and in particular, from beer.
Table 3. Adsorption of purines and close compounds.
| Target |
Matrix |
Adsorbent |
| Uric acid |
Aqueous solutions |
2-hydroxyethyl methacrylate and ethyleneglycol dimethacrylate in the shape of granules |
| Uric acid |
Aqueous solutions |
Zinc oxide nanoparticles loaded on activated carbon |
| Uric acid |
Aqueous solutions |
Pitch-based spherical activated carbon (PSAC) modified by CVD of NH3 |
| Uric acid |
Aqueous solutions |
Polyethyleneimine/SiO2 |
| DNA and RNA |
- |
Activated carbon coated with polyvinylpyrrolidone, dextran or coconut flours |
| DNA nucleobases (guanine, adenine, cytosine and thymine) |
- |
Graphene |
| DNA nucleobases (guanine, adenine, cytosine and thymine) |
- |
Carbon nanotubes, dendrimers and graphene |
| DNA nucleobases (guanine, adenine, cytosine and thymine) |
Aqueous solutions |
Graphene |
| Adenine, adenosine and AMP |
Aqueous solutions and beer |
Activated carbon derived from beer lees |
| DNA nucleobases (guanine, adenine, cytosine and thymine) |
- |
Graphene |
| DNA nucleobases (guanine, adenine, cytosine and thymine) |
- |
Graphene |
| Adenine, thymine and radicals |
|
Single-wall carbon nanotubes |
| Adenosine, guanosine and guanine |
Wort, beer and low-malt beers |
Activated charcoal |
| DNA nucleobases (guanine, adenine, cytosine and thymine) |
|
Graphite |
| Nucleotides and nucleosides |
|
Activated carbon |
| Adenine and xanthine |
Aqueous solutions |
Dextran T40 and Sephadex G-10 |