Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Gour Mohan Das | + 5565 word(s) | 5565 | 2021-10-20 03:46:08 | | | |
| 2 | Bruce Ren | Meta information modification | 5565 | 2021-11-01 02:07:49 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Das, G.M. Biosensing Using SERS Active Gold Nanostructures. Encyclopedia. Available online: https://encyclopedia.pub/entry/15546 (accessed on 07 February 2026).
Das GM. Biosensing Using SERS Active Gold Nanostructures. Encyclopedia. Available at: https://encyclopedia.pub/entry/15546. Accessed February 07, 2026.
Das, Gour Mohan. "Biosensing Using SERS Active Gold Nanostructures" Encyclopedia, https://encyclopedia.pub/entry/15546 (accessed February 07, 2026).
Das, G.M. (2021, October 29). Biosensing Using SERS Active Gold Nanostructures. In Encyclopedia. https://encyclopedia.pub/entry/15546
Das, Gour Mohan. "Biosensing Using SERS Active Gold Nanostructures." Encyclopedia. Web. 29 October, 2021.
Copy Citation
Surface-enhanced Raman spectroscopy (SERS) has become a powerful tool for biosensing applications owing to its fingerprint recognition, high sensitivity, multiplex detection, and biocompatibility. This review provides an overview of the most significant aspects of SERS for biomedical and biosensing applications.
SERS
biosensing
gold nanoparticles
1. Introduction
Surface enhanced Raman scattering (SERS) can be defined as amplified Raman scattering by the presence of plasmonic nanostructures (generally metallic nanoparticles) in the proximity of the analyte molecules[1]. Upon excitation with appropriate light interacting with the sub-nanometer metallic structures, the collective oscillation of conduction electrons is observed, generating an ultra-strong electromagnetic (EM) near-field in the proximity of the nanostructure surface. This phenomenon is known as localized surface plasmon resonances (LSPRs) and it is at the foundation of SERS spectroscopy. SERS spectroscopy has particular advantages for biosensing applications: (i) it allows reconstruction of a detailed spectral profile providing all the structural, compositional, and conformational features of the analyte molecules; (ii) it enables multiplex detection with single wavelength excitation, therefore several molecules can be monitored at the same time; (iii) it is not destructive; (iv) it allows measurements in biological fluids since the water spectrum is rather weak; (v) SERS is resistant to photo-bleaching and photo-degradation compared with fluorescence and is suitable for long-term monitoring, and (vi) SERS does not require high sample concentrations. In other words, SERS spectroscopy combines the intrinsic advantages of Raman spectroscopy with high sensitivity. Indeed, the million-fold enhancement enables SERS detection down to a single molecule level[2][3].
The key element of SERS spectroscopy is the realization of the metallic nano-substrate. The development of several bottom-up chemical synthesis procedures or advanced top-down nanofabrication technologies allows for obtaining a variety of nanostructures with tunable geometries. Indeed, SERS-active nanostructures can be designed with different sizes, shapes, compositions, aggregations, and coatings, allowing the possibility of response tuning for specific detection purposes. The SERS substrate can be optimized for the near-infrared excitation laser, thus avoiding auto-fluorescence from biological samples and minimizing their photodamage. Within the group of noble metal nanostructures, gold-based nanostructures or gold nanoparticles (AuNPs) are mostly used for biosensing applications due to their biocompatibility, tunability, their unique optical and electronic properties, together with their nontoxic nature when compared to other nanomaterials, such as metal oxides or carbon-based nanomaterials in general[4][5][6]. As a result, there have been several impressive developments and applications of SERS active gold nanostructures for biosensing applications[7][8][9].
2. SERS Substrates Used for Biosensing
At the core of SERS biosensing is the interaction between the analyte molecule and the selected substrate. Therefore, the selection/fabrication of the SERS substrate is a key element to determine the performance of the sensor. The first SERS substrate was fabricated using the electrochemically roughened metal electrodes, and the main limitations are the reduced control on plasmon resonance as well as the hotspot due to the aggregation with low tenability, even though one can produce a large-area SERS substrate using this technique[10]. Due to the development of nanotechnologies, the use of metal nanoparticles as a highly sensitive and more controlled SERS substrate for biosensing has increased exponentially. Gold and silver are the most commonly used materials for the fabrication of SERS active substrates for enhancing the Raman scattering signal in the visible region. The fundamental mode of the LSPR band in the visible region depends upon the dielectric constant of gold and silver. Even though silver has the largest plasmon resonance in an easily accessible spectral region, due to its strong toxicity to living systems, Ag is seldom used for in vivo analysis, but it is good for in vitro ultrasensitive detection [11][12]. Whereas the Au provides more chemical stability, Au nanostructures are good for excitation in visible and near-infrared regions and are commonly used in intracellular or in vivo studies due to their excellent biocompatibility[13][14].
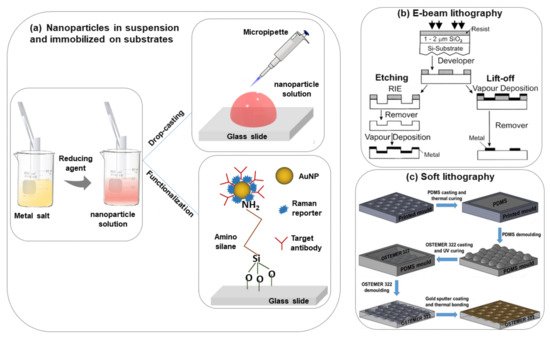
The most commonly used nanoparticle synthesis techniques are the wet chemistry method and the laser ablation technique[15][16]. Spherical metallic colloidal nanoparticles generally used in SERS experiments can be synthesized by reduction of metal salts using the wet chemistry method[17][18] (see Figure 1a). Nanoparticles can be alternatively synthesized using laser ablation[16]. A metal target is placed at the bottom of a solution and a pulsed laser is nearly focused at its surface; the heating and photoionization processes cause the metal to change the state of aggregation, forming liquid drops, vapors, or a plasma plume. Even though this method is less easy than the previous one, using this technique one can still fabricate the plasmon active substrate without capping agents, which allows an easier functionalization of nanoparticles[19].
The size and shape of the nanoparticles strongly influence the performances of the substrate for sensing applications. If the size of the metal nanoparticles on the SERS substrate is too large then the multipole (non-radiative modes) excitation occurs rather than a dipole. Due to this, the overall enhancement in the Raman scattering will reduce. Based on the literature, nanoparticles that have a size between 10–100 nm are more suitable for SERS experiments[22]. The shape of the nanoparticle is an additional key parameter to consider for improving the SERS performances in metal nanoparticle suspensions. Besides nanospheres, many other shapes, including nanorods, nanoprisms, nanocubes, nanoplates, nanostars, etc. have been proposed and demonstrated for different biosensing applications[23][24]. Additionally, core-shell nanoparticles, alloy nanoparticles, core-satellite nanoparticles, and yolk-shell and core molecule-shell have been utilized in SERS investigations to improve sensor performance[25][26][27][28]. Some popular shapes of metal nanostructures that are used very often for different biosensing experiments have been shown in Table 1. The shape and size of colloidal metal nanoparticles always have a strong influence on the EF of the SERS signal[29]. The obtained EF is summarized in Table 1, but a fair comparison between different shapes is complicated by the use of different analyte molecules.
Table 1. Different shapes colloidal SERS active AuNPs.
| Shape | Dimension | Detected Molecule | Laser | EF | Ref. |
|---|---|---|---|---|---|
| Nanoflower | 400 nm | Rhodamine 6G | 785 nm | 105 | [[32]] |
| 45 nm | Rhodamine 6G | 532 nm | 106 | [[30]] | |
| Nanostar | 105 nm | Crystal violet | 785 nm | 107 | [[31]] |
| 130 nm | 4-mercaptobenzoic acid (MBA) | 785 nm | 109 | [[32]] | |
| Nano-bowtie | Height: 40 nm, Gap: 8 nm, Edge: 90 nm |
Trinitrotoluene (TNT) | 785 nm | 105 | [[33]] |
| Height: 25 nm, Gap: 8 nm, Edge: 100 nm |
Bi-(4-pyridyl) ethylene (BPE) | 785 nm | 107 | [[34]] | |
| Nanorod | Length: 69 nm, Width: 24 nm | Rhodamine 6G | 532 nm | 106 | [[35]] |
| Length: 41 nm, Width: 18 nm | Rhodamine 6G | 633 nm | 107 | [[36]] | |
| Nanocube | Edge length: 170 nm | Rhodamine 6G | 633 nm | 105 | [[37]] |
| 32–72 nm | Crystal violet | 785 nm | 106 | [[38]] | |
| Edge length: 84 nm diameter 55 nm |
1,8-octanedithiol (C8DT) | 785 nm | 1010 | [[39]] | |
| Nanosphere | Size range: 15–40 nm | Crystal violet | 633 nm | 103–104 | [[40]] |
| 36.5 ± 6.3 nm | 4 ATP | 785 nm | 105 | [[41]] | |
| Nanospheroid | semi-major axis: 55 nm, semi-minor axis: 30 nm |
Bi-(4-pyridyl) ethylene (BPE) | 633 nm | 105 | [[42]] |
| 50 nm | Crystal violet | 514 nm | 108 | [[43]] | |
| Nanoshell | Core: 46 nm, Shell: 22 nm | Thiobenzoic acid (TBA) | 785 nm | 105 | [[44]] |
| Hollow nanoshell | Diameter 250 nm | Rhodamine 6G | 785 nm | 105 | [[45]] |
| Diameter 233 ± 14 nm | Methylene blue | 532 nm | 105 | [[46]] |
In metal nanoparticle suspensions for SERS sensing applications, it is important to control the nanoparticle aggregation and distribution. The colloidal nanoparticle solution can be directly spotted on a glass slide followed by evaporation of the solution (drop-casting)[46][47] (Figure 1a). This method is easy and fast but without specific control of the nanoparticle distributions and signal reproducibility. However, to obtain an ordered nanoparticle distribution over a slide, typically a glass surface, the silanization protocol with (3-Aminopropyl) trimethoxysilane (APTMS) can be applied[47] (Figure 1a). In this case, a strong interaction between –NH2 groups and the nanoparticles occurs which helps to prevent spontaneous coalescence (it is observed in simple drop-casting on the glass substrate) and this protocol is valid for any type of dielectric substrate like glass, ITO, silicon, etc.[48][49]
Alternative strategies for in-liquid SERS detection are currently based on chemically-driven aggregation or optical trapping of metal nanoparticles in the presence of the target molecules[50]. An interesting example is the use of contactless manipulation methods, like laser tweezers, where the optical forces are used. A nanoparticle is subjected to two forces: (i) A gradient force that is attractive towards the high-intensity region of the laser beam if the excitation wavelength is longer than the surface plasmon resonance of the nanoparticle and repulsive in the opposite case; (ii) a radiation pressure force that propels the nanoparticle along the propagation direction of the beam[51][52].
In the last few years, researchers have developed different types of metallic nanostructures as SERS-active substrates via advanced nanofabrication techniques[20][21][53][54][55][56][57][58]. Electron beam lithography is one of the most popular techniques used for the fabrication of the SERS substrate (shown in Figure 1b), where an electron beam of 10–30 keV incident on the silicon wafer (covered by either positive and negative resist). After the etching using an electron beam, there are two ways to fabricate the SERS substrate. One process consists of the chemical etching that follows the electron beam exposing, the dissolution of the remaining resist layer, and deposition of metal, and finally, the substrate is covered with metal. Another technique involves metal deposition immediately after the electron beam exposition. After removal of the photo-resist layer, the substrate will present a series of isolated nanoparticles, separated by regions where only bare Si substrate is exposed[20]. The main advantage of this process is the possibility to control the size, shape, and inter-particle distance between the nanoparticles. In addition, with the electron beam lithography, optical lithography has been also used for the fabrication of SERS active substrates[53]. Different SERS active substrates fabricated by lithography techniques are shown in Figure 2. Generally, all these techniques provide several advantages in terms of substrate reproducibility and sensitivity but require serial on-surface fabrication steps that are error-prone and time-consuming. Furthermore, costly instrumentation and procedures, including the use of a cleanroom facility, are required and it is very difficult to cover the large area of SERS devices using these approaches.
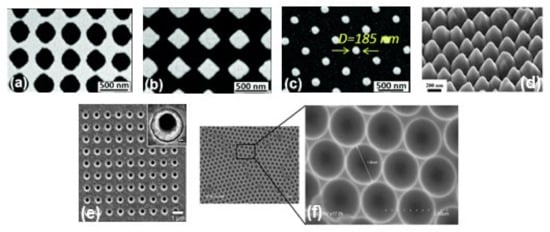
Figure 2. SEM images of metallic periodic nanostructures nanohole arrays (a), diamond-shaped nanoparticle arrays (b), arrays of cylindrical nanoparticles (c) fabricated by different development times 95 s, 110 s, and 165 s, respectively. Reproduced from ref.[59] with permission from the Royal Society of Chemistry. (d) 45° tilt view of nanostructure arrays with Au coating thickness 60 nm. Reproduced with permission from Springer Nature Publications of ref.[35]. (e) nanohole array with 500 nm diameter, the inset shows the magnified image of one of the holes. Reprinted (adapted) with permission from ref.[53], Copyright 2010 American Chemical Society. (f) the cavity array substrate of 2 μm fabricated applying polystyrene microsphere template approach[21].
Recently, it has been shown that processes based on self-assembly represent an easier, quicker, and cheaper bottom-up option than nanolithography for the fabrication of regular micro-and nanopatterns[57][58][59] (see Figure 1c). Indeed, several SERS sensing devices have been fabricated using the nanosphere lithography technique. Cusano et al. demonstrated that polystyrene spheres (with diameters ranging from 200 to 1000 nm) self-assembled at the air-water interface into ordered hexagonal close-packed arrays and covered by a gold conformal layer can work as an efficient SERS substrate (basic close-packed array, CPA, monolayer)[58]. Their proposed fabrication procedure was inexpensive, simple to implement, and well-ordered. The substrates can be deposited on a large area, on any type of surface including the fiber tip, and can be efficiently exploited to create optical fiber SERS probes[60].
3. Biosensing Using SERS
Label-free SERS detection, that is the measurement of the intrinsic enhanced Raman signature of the analyte molecule, or indirect SERS based-sensor has been used for the detection of different complex molecules for applications in biomedicine[61] including nucleic acids, proteins, and cells. In this section, we summarize a few cutting-edge recent reports related to SERS-based biomolecule detection using gold nanoparticles/nanostructures, as reported in the Table 2.
3.1. Sensing of DNA/RNA
In modern medicine, the identification of DNA and RNA has significant implications for the analysis, diagnosis, and treatment of genetic and infectious diseases. Moreover, the recent global SARS-CoV-2 pandemic, with the emergence of the COVID-19 and its multiple variants, illustrates the unmet need for rapid, flexible nucleic acid testing technology for disease diagnostics[62]. To date, RT-PCR is the gold standard oligonucleotide detection technique, which however requires several reagents and procedural steps (e.g., nucleic acid isolation and retro-transcription), expensive equipment, reagent costs, good laboratory practices, and skilled personnel[63]. Indeed, the basic RT-PCR assay relies on extraction and purification of the nucleic acid, then exponential amplification of the target sequence, using a thermostable polymerase enzyme and specific primers[63]. An important issue is also the time for the analysis, which goes from a few hours to 1–2 days. The needs for designing specific primers for the target require handling by experienced operators, capable of detecting errors. The technique, if not performed in rigorous conditions and with controlled reagents, is particularly prone to false-positive results. In this matter, SERS is one of the useful promising ultrasensitive techniques to accurately identify specific DNA/RNA fragments[64][65][66][67].
Wang et al.[64] synthesized gold plasmonic nanopores by in situ reductions of gold on the confined tip of a glass nanopipette, where a bias potential was applied to drive DNA oligonucleotides and amino acids translocating through the nanopores (where the hot spots are located), which enables the collection of rich structural information for biomolecules by SERS technique (see Figure 3a,b). With the application of the bias potential, the Adenine SERS signal can be monitored over a large range of concentrations from 10−4 to 10−9 M (Figure 3c). The SERS spectra of four nucleobases of A, T, G, and C display a clear fingerprint of Raman features that can be useful for the direct identification of single nucleobases in DNA oligonucleotides (Figure 3d). Indeed, the authors demonstrated that the SERS-based nanopore detection can directly provide structural information, which enables it to distinguish DNAs with a single nucleobase difference with high spatial resolution and high sensitivity.
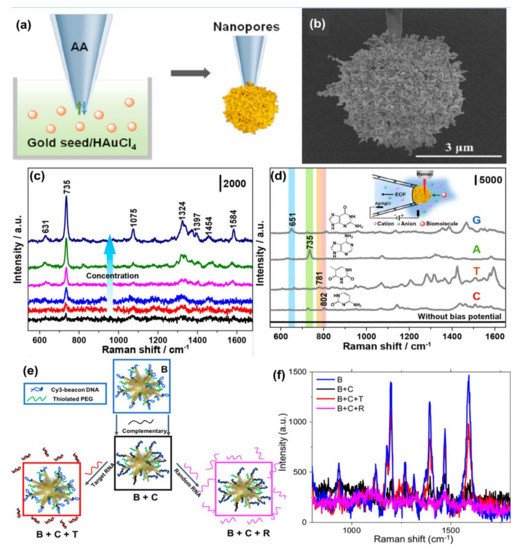
Figure 3. Panel (a) is the schematic of the synthesis of plasmonic nanopores process. SEM image of the gold plasmonic nanopores is shown in panel (b). Panel (c) represents SERS spectra of A translocating through gold plasmonic nanopores with a bias potential of −1 V, from low to high 10−9 to 10−4 M, respectively. Panel (d) represents the SERS-based nonresonant molecules detection of the four nucleobases (G, A, T, and C). The scheme of measurement setup (inset). Reprinted (adapted) with permission from ref.[64], Copyright 2019 American Chemical Society. Panel (e) shows the Au nanostars are functionalized with a Cy3-tagged beacon DNA, creating the SERS-active nanostar probes (B), and then hybridized with a complementary oligonucleotide (B + C). Upon exposure to the viral RNA targets, the complementary oligonucleotide dehybridizes from the beacon and hybridizes with the viral RNA (B + C + T), returning the beacon to its original hairpin conformation and leading to SERS signal recovery (B + C + R), Panel (f) is the SERS signal “ON-OFF-ON” switching: ON with beacon in hairpin conformation (B); OFF when the beacon is hybridized with 500 nM complementary oligo (B + C); then ON again upon exposure of SANSPs to 500 nM target viral RNA (B + C + T); Signal recovery was not observed after a random RNA sequence (500 nM) was introduced (B + C + R). Reprinted (adapted) with permission from ref.[68], Copyright 2020 American Chemical Society.
Recently, Kim et al.[65] developed a SERS-based plasmonic biosensor for the label-free multiplex detection of miRNAs (miR-10b, miR-21, and miR-373), which are relevant to cancer metastasis. The SERS-based nanoplasmonic biosensor was fabricated with the application of a head-flocked gold nanopillar substrate and a complementary DNA probe platform was used for selective targeting of miRNAs with an extremely low detection limit, low signal fluctuations, and high signal stability. The label-free SERS nanoplasmonic biosensor was used to discriminate between target miRNAs (miR-10b, miR-21, and miR-373) by performing the SERS signal measurement according to the hybridization process with ultra-high sensitivity (LODs of 3.53 fM, 2.17 fM, and 2.16 fM, respectively). According to the report, this plasmonic biosensor offers many advantages such as label-free detection, high selectivity, high sensitivity, and multiplex detection capability.
Several indirect sensing methods have been reported in the literature for improving the detection of oligonucleotides using SERS. In most of the SERS-based DNA/RNA sensors, a Raman tag is attached with the plasmonic nanostructures, and indirect highly sensitive, and highly reproducible sensing is performed. Indeed, sandwiched DNA/RNA structures where the movement of the Raman reporter or organic dye close to or away from the plasmonic nanostructure is realized by standard hybridization leading to a double-stranded DNA (or DNA/RNA hybridization) or by the formation or decomposition of a hairpin chain (stem-loop configuration) has been demonstrated by several groups[66][67]. The use of indirect approaches produces significantly high SERS signals for the detection of DNA with a notably low concentration (<10 fM), high sensitivity, specificity, and reproducibility[67]. The combination of the use of high-performing SERS substrates and specific DNA targets with Raman dyes can enhance the multiplexed detection capability of the SERS sensor and crucially, provide quantitative information[69]. In a recent paper, Fabris et al.[68] demonstrated the use of the SERS approach for intracellular monitoring of RNA mutations in the influenza A virus. Gold nanostars, functionalized with a DNA hairpin structure decorated with a Raman-dye was designed to selectively extend/fold in the absence/presence of the viral RNA targets. “OFF-ON” switching of the SERS signal was observed when the fluorophore was away from or close to the gold nanostar surface (Figure 5e,f). The degree of signal recovery correlated with the number of genetic mutations has been reported. The authors for the first time employed molecular beacon-based SERS probes to detect viral RNA targets in intact individual cells. Indeed, the experiments carried out with HeLa cells transfected with plasmids coding for either hemagglutinin (HA) segment or two other segments (negative controls) demonstrate the functionality of the molecular beacon-based SERS probes in intact individual cells with high sequence sensitivity. These results suggest the applicability of these probes for multiplexed detection and quantification of viral RNAs in individual cells, including the RNA from COVID-19, with an approach that can account for the viral population diversity.
3.2. Sensing of Proteins
Detection and quantification of proteins in biological fluids associated with specific diseases at the point-of-care (POC) have attracted increasing interest for many diverse medical applications. State-of-the-art immunological methods require direct fluorophore labeling of the protein, labeling secondary reagents that bind to the protein, or labeling secondary reagents that bind to a tag such as biotin. These methods can be carried out with ELISA equipment making it inexpensive and readily available. However, the use of labels can reduce activity or selective binding, labeled secondary reagents are not always available, there is a limit on the number of proteins that can be detected at the same time as well as the photo-bleaching and degradation of the fluorophore. Due to these issues, there is a push in the research community to use label-free methods, for instance, SERS spectroscopy for protein detection. Obtaining SERS data from solutions of proteins is challenging as their structure is bigger than molecules, can easily denature, aggregate, modify their conformation generally present in complex matrices (biological fluids, blood, urine, cells, etc.) at low concentrations[70].
One direct sensing protocol using the concept of in-liquid molecular detection via SERS has been reported by Fazio et al. In this work, the authors have implemented a label-free, all-optical SERS sensor for the detection of biomolecular in buffer solution (Phenylalanine (Phe), Bovine Serum Albumin (BSA) and Lysozyme (Lys) at concentrations down to few μg/mL) which exploits the radiation pressure to push gold nanorods on a surface and form SERS-active aggregates in buffered solutions of amino acids and proteins. The schematic of the principle of SERS detection of biomolecules using optical force has been shown in Figure 4a, also the SERS detection of BSA molecule in phosphate buffer solution has been shown in Figure 4b. Here, the estimated maximum enhancement factor was reported around 105, using the excitation laser 632.8 nm. According to the authors, the main advantages of this sensor are that the detection happens in the natural environment of biomolecules, the sensing is rapid, the technique is experimentally simple, reliable, and intrinsically scalable to lab-on-chip devices[50].
Brule et al., reported the study of bovine serum albumin (BSA) protein conformations using SERS, where they used self-assembly of raspberry-like AuNPs immobilized for the fabrication of SERS substrate[71]. The substrate is excited by a 784 nm laser and the estimated maximum enhancement factor reported is around 107 and the limit of detection is almost 10 pM. Also, the dynamic SERS record enables discriminating the physisorption and the chemisorption events using multivariate analysis. This event is confirmed by different characteristic SERS signals recorded from the hydrophobic amino acid fingerprints, like tryptophan, tyrosine, leucine, and histidine. According to the authors, tryptophan is a specific biomarker for the unfolded conformation of the BSA. Indeed, BSA has only two tryptophans in its structure, one is at the protein surface and the other one is located in the hydrophobic core. During BSA unfolding, the tryptophan in the hydrophobic core is exposed and is free to give a strong and specific signal, thanks to the high Raman vibrational activity of the benzoic structure. Instead, the tryptophan at the protein surface has the benzoic ring still blocked by the surrounding amino acids and unable to give vibrational signals. For these reasons, tryptophan-associated bands are considered good markers of BSA unfolding. Moreover, when BSA is unfolded also other amino acids, such as histidine, that surround the inner tryptophan are exposed and give Raman signals developing a high electrostatic affinity with gold[71].
Another indirect protein sensing platform via SERS has been reported by Zengin et al., where they present a homogeneous detection method utilizing monoclonal anti-tau functionalized hybrid magnetic nanoparticle (MNP) probes and polyclonal anti-tau immobilized AuNPs as SERS tags in solution[72]. The tau specificity on BSA and immunoglobulin G (IgG) was also tested using this technique. The schematic of the experimental procedure and the SERS spectra of monoclonal anti-tau functionalized hybrid nanoparticles exposed to BSA (500 nM), IgG (500 nM), tau (500 nM), and a solution with equal amounts (500 nM) of BSA, IgG, and tau has been shown in Figure 4c,d. According to the authors, this SERS-based sandwich assay system possesses advantages over the current methods such as ELISA, LSPR, and chromatography which contain label-free and rapid detection using a simple and cost-effective substrate fabrication technique. This approach might provide accurate, sensitive, and more selective detection of tau than current protein detection methods with the lowest limit of detection for tau solution of below 25 fM, which is comparable to the sensitivity of conventional optical biosensing methods[72].
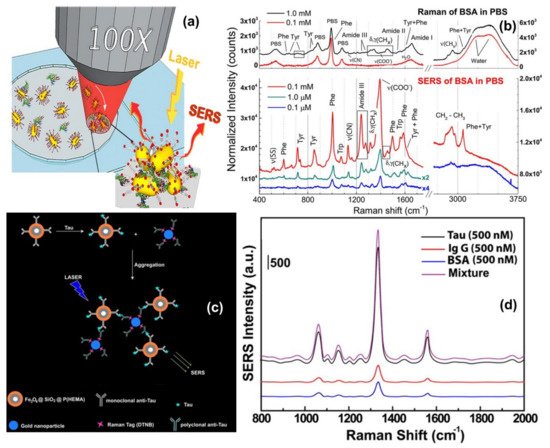
Figure 4. Panel (a) shows the schematic of the principle of SERS detection of biomolecules using optical force. Panel (b) shows the comparison between Raman spectra and SERS spectra of BSA and Phenylalanine in phosphate buffer solution. Reproduced with permission from Springer Nature Publications of ref.[50]. Panel (c,d) are the schematic of the experimental procedure of indirect protein sensing platform and the SERS spectra of monoclonal anti-tau functionalized hybrid nanoparticles exposed to BSA (500 nM), IgG (500 nM), tau (500 nM), and a solution with equal amounts (500 nM) of BSA, IgG, and tau. Reprinted (adapted) with permission from ref.[72], Copyright 2013 American Chemical Society.
3.3. Sensing of Cells
Label-free SERS-based methods can be used to detect intrinsic spectroscopic signatures from living cells that can be acquired to identify different cell components. Bando et al.[73] used 3D SERS-imaging to track AuNPs moving in the cytosol of living cells. The simultaneous detection of the SERS signal and spatial position of the nanoparticles enabled to reconstruction large amount of information on biomolecular events. Time-space-spectrum variable analysis allowed the visualization of molecular dynamics such as dissociation of proteins in biological reactions or the study of molecules associated with the transportation process such as vesicle transport, nuclear entry, and protein diffusion.
SERS fingerprinting of individual molecules enables the possibility of cancer screening. Liu et al.[74] proposed the use of economic, storable, biodegradable, and easy fabricated paper-based SERS substrate for automated, rapid, and non-invasive cancer cell screening. Different and reproducible SERS spectra were detected from normal and cancerous cells due to specific biomolecular changes in cancerous cells (shown in Figure 5a,b). A diagnostic algorithm based on band ratio analysis allowed the discrimination with a sensitivity and specificity up to 70%.
Since direct SERS-analysis and full band assignment suffer from SERS signal stability and reproducibility, SERS tags for indirect cancer detection have been additionally proposed. Wu et al. developed three SERS-active AuNPs with different shapes (nanosphere, nanorods, and nanostars) modified with a Raman reporter molecule (4-mercaptobenzoic acid, 4-MBA) for detection of circulating tumor cells (CTCs) from blood. These nanoparticles showed a strong SERS signal and excellent sensitivity due to the Raman reporter (LOD of 1 cell/mL) and high specificity due to conjugation of the targeted ligand (acid folic) which can reduce the nonspecific catching or uptake by healthy cells and can recognize CTCs of various cancers overexpressing of folate receptor alpha (including ovarian, brain, kidney, breast, lung, cervical, and nasopharyngeal cancers).
Cancer cells are characterized by specific bioreceptors present in their plasma membranes. The detection and accurate identification of bioreceptor expression on the cell surface represents a crucial step in the diagnostic process. In this scenario, the high multiplexing capability of SERS has the potential to provide accurate molecular phenotyping of the individual cancer cells. Bodelón et al.[75] proposed a new class of SERS tags based on poly (N-isopropylacrylamide) (pNIPAM) encapsulated AuNPs for the multiplex detection of tumor-associated protein biomarkers. The authors demonstrated that this system overcame particle aggregation issues, allowed them to achieve high resonance Raman scattering enhancements leading to reproducible high-intensity signals, and allowed multiplex detection and imaging of three important tumor-associated biomarkers (EGFR, EpCAM, or CD44), for discrimination of A431 tumor and 3T3 2.2 nontumor cells with a single excitation laser line.
Real-time monitoring of a single cancer cell, under different conditions, and extracellular stimuli, can be assessed by coupling SERS spectroscopy with single-cell microfluidics as reported by Willner et al.[76]. A single prostate cancer cell was trapped thanks to the microfluidic device in a droplet and the developed wheat germ agglutin-functionalized SERS nanoprobes used for spectroscopic identification of glycans on the cell membrane. The analysis was performed in two steps. First, a large sample area was scanned with a low spectral resolution enabling the ‘fast’ identification of SERS regions of interest in cancer cells. In a second step, the SERS signal from WGA-modified metallic nanoparticles was used to ‘slower’ monitor the expression of glycans at a higher spectral resolution and to screen the dynamic cell-to-cell variability in a higher throughput fashion.
Intrinsic SERS signals can be used to detect specific biomarkers or physiologically relevant biomolecular species inside or near individual cells, such as important metabolites in normal and tumor cells, or other cellular components such as exosomes. The application of SERS to identify the presence of extracellular metabolites, secreted by cancer cells and relevant to tumor biology, including tryptophan, kynurenine, and purine derivatives, has been demonstrated by Plou et al.[77]. The proposed SERS-based strategy used a plasmon active substrate comprising a superlattice of Au nanoparticles, as the source of enhancement for the Raman signal from the analytes (for details see in Figure 5c–e). The authors claimed that the sensitive and cost-effective plasmonic substrate was effectively combined with 3D cell culture models, which more closely recreate the biochemical and biophysical factors in the tumor microenvironment, toward real-time imaging of heterogeneous metabolic alterations and cytotoxic effects on tumor cells, which will have significance in diagnosis and therapy.
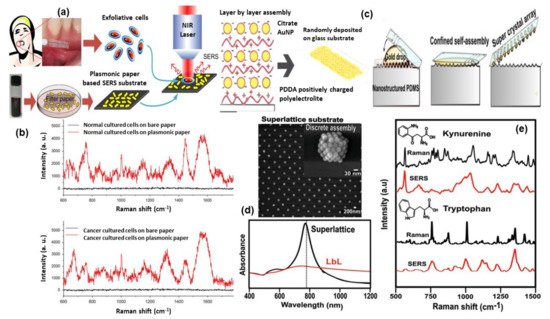
Figure 5. Panel (a) shows the illustration of paper-based SERS substrate and SERS examination of the exfoliated cells obtained from oral-cancer patients. Panel (b) represents the SERS spectra from the exfoliated cells: normal tissue (top) and cancerous tissue (down)[74]. Panel (c) shows the methodology of super-lattice SERS substrate fabrication and its SEM image and Vis-NIR spectra are shown in panel (d). Panel (e) shows the comparison of Raman and SERS spectra for kynurenine (Kyn) and tryptophan (Trp). Reproduced with permission from Wiley publications of ref.[77].
Managò et al., demonstrated one interesting way for SERS probing of red blood and leukemic cells (see Figure 6a–d). In that experiment, diatom frustules have been used which is formed by a complex, intricate but regular dielectric 3D nanostructure. The diatom frustules properly metalized trigger broadband LSPR in the red and infrared spectral range and can be used as SERS substrate for cell probing. The SERS spectra from red blood cells and leukemia cells were mainly related to the cell membranes whose impairment is responsible for several pathologies. This allowed the limitations of conventional spontaneous Raman spectroscopy, where the scattered light comes from the cell as a whole, to be overcome[78].
Quantitative and label-free detection of glycerophosphoinositol (GroPIns) using SERS techniques has been reported by De Luca et al., GroPIns are cell metabolites regulating important cell biological functions. Indeed, their increased concentration in the cell cytosol has been associated with several diseases, including thyroid cancer[79]. The detection of GroPIns cellular levels can be considered a biochemical marker of photo/physiological conditions. In their paper De Luca et al., used Au fishnet nanostructures, fabricated using e-beam lithography, as SERS active substrate for sensitive detection of GroPIns in complex matrices. The authors demonstrated that the proposed SERS substrate provided more reproducibility as compared with colloidal nanoparticles[79] together with a good enhancement. Indeed, the proposed SERS-based approach allowed for a rapid (acquisition time: 1 s), quantitative (accuracy: 6%), and a sensitive (detection limit: 200 nM) detection of the GroPIns in complex matrices eliminating the need for labels/dyes, long sample preparation and the use of large volume samples (single-cell volumes can be used).
Tian et al., recently developed a SERS sensor for the detection of exosomes, small extracellular vesicles (30–150 nm) which transfer and deliver encapsulated molecules from their originating cells, as a cancer biomarker. In the proposed indirect SERS sensor, 4-mercaptobenzoic acid (4-MBA) embedded in Au overcoated nanostars (AuNS@Au) was used as a Raman reporter, bivalent cholesterol labeled DNA anchor conjugated onto AuNS@4-MBA@Au formed SERS nanoprobes and exosome-bound magnetic beads were used as the capture probes. Exosomes are specifically captured by immune magnetic beads, and then SERS nanoprobes are fixed on the surface of exosomes by hydrophobic interactions between cholesterol and lipid membranes, thus forming a sandwich-type immune complex. The immune complex can be magnetically captured and produce enhanced SERS signals. In the absence of exosomes, the sandwich-type immune complex cannot be formed, and thus negligible SERS signals are detected. The degree of immune-complex assembly and the corresponding SERS signals are positively correlated with the exosome concentration over a wide linear range of 40 to 4 × 107 particles per µL and the limit of detection is as low as 27 particles per µL[80].
SERS-based sensors can be used to monitor certain physiological processes as well as the real-time release of drugs in living cells[81]. Managò et al., recently demonstrated the use of hybrid nanoparticles, consisting of diatomite nanoparticles decorated with gold nanospheres and covered by a pH-sensitive gelatin shell, for label-free imaging of the nanovector localization and local SERS-sensing of the intracellular release of galunisertib in living colorectal cancer cells (see Figure 6e–g). AuNPs strongly enhance the SERS signal of the drug-loaded in the diatomite pores allowing the tracing and quantification of its release in living cells over days with high sensitivity (down to sub-femtogram of the drug). Furthermore, the encapsulation of galunisertib in the nanoplatform can help to lower the amount of drug required to inhibit the metastatic process in cancer cells, reducing the formation of drug-related toxic metabolites.
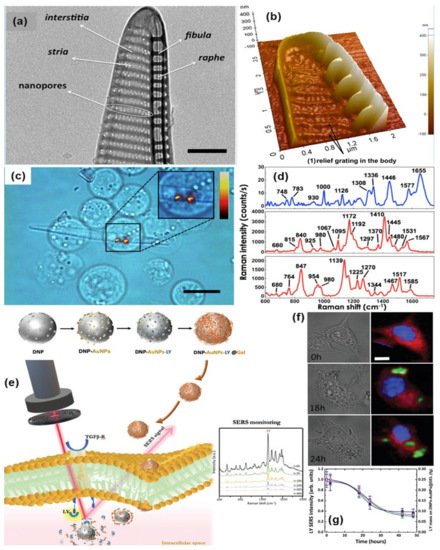
Figure 8. TEM and AFM image of a P. multistriata single valve is shown in panels (a,b). Panel (c,d) shows the optical image of REH LCs over a single metalized diatom valve and the conventional Raman spectrum of a single REH cell. Reprinted (adapted) with permission from ref.[78], Copyright 2018 American Chemical Society. The schematic diagram of hybrid nanosystem (diatomite nanoparticles-AuNPs-LY@Gel) synthesis and internalization in colorectal cancer (CRC) cells has been shown in panel (e). Panel (f,g) show optical image and Raman mapping images showing the internalization of DNP-AuNPs-LY@Gel (50 μg mL−1) into CRC cells after 0, 18, and 24 h of incubation (scale bar = 10 μm) and time-dependent LY SERS signal from the hybrid nanocomplex in living CRC cells. Reproduced with permission from Wiley publications of ref.[81].
Table 2. A brief summary on biosensing using SERS.
| Type of Nanoparticles/ Substrate |
Target | Type | Detection Mechanism |
Enhancement Factor (EF) or Limit of Detection (LOD) |
Ref. |
|---|---|---|---|---|---|
| Au nanopores | DNA | Direct | Bias potential + SERS detection of DNA | EF: 106 | [[64]] |
| Au nanopillar | miRNA | Direct | DNA/RNA hybridization + SERS detection of DNA/RNA |
LOD: 3.53 fM | [[65]] |
| Au NPs-decorated silicon nanowire array |
DNA | Indirect | stem-loop DNA/target DNA hybridization + SERS detection of dye molecule |
EF: 7.24 × 105 LOD: 10 fM |
[[67]] |
| Spherical Au nanoparticles |
miRNA | Indirect | Symmetric signal amplification + SERS detection of Cy3 | LOD: 7.5 fM | [[69]] |
| Au nanostar | RNA | Indirect | Identify and quantify RNA mutations through SERS | / | [[68]] |
| Au nanorod | BSA | Direct | Optical Tweezers + SERS detection of BSA | EF: 105 | [[50]] |
| Raspberry-like assembled Au nanoparticles |
BSA | Direct | Dynamic SERS of BSA | EF: 104–107 LOD: 10 pM |
[[71]] |
| Au NPs + magnetic NPs | tau protein | Indirect | SERS-based sandwich assay | LOD: 25 fM | [[72]] |
| Au NPs | Hela cells | Direct | Intracellular detection of proteins/cytosol by SERS |
/ | [[73]] |
| Au nanorods | Oral cancer cell | Direct | Cancer cell screening using SERS | Sensitivity: 70% Specificity: 60% |
[[74]] |
| Au nanosphere, rods, stars | Circulating tumor cells |
Indirect | SERS detection of Raman reporter (4-MBA) for identification of cancer cells |
EF: 104 | [] |
| Au octahedral NPs | Tumor Cells | Indirect | Detection and imaging of cancer cells by SERS tags | / | [[75]] |
| Functionalized Au NPs | Prostate cancer cell | Indirect | Imaging and identification of Glycans in cell membrane by detection of the SERS probe | / | [[76]] |
| Au superlattices | tumor metabolites |
Direct | Identification of cell metabolites by SERS on a chip device | / | [[77]] |
| Au metallized diatom | red blood, leukemic cells |
Direct | SERS detection of cell membrane | EF: 106 | [[78]] |
| Au fishnet | Cell metabolites |
Direct | SERS detection of Glicerophosphoinositol |
LOD: 200 nM | [[79]] |
| Au nanostars | Exosomes | Indirect | SERS-based sandwich assay | LOD: 4 × 104 particles per µL |
[[80]] |
| Diatomite nanoparticles decorated with Au NPs |
Drugs in colorectal cancer cells |
Direct | SERS-detection of Drugs in living cells | EF: 105 | [[81]] |
References
- Judith Langer; Dorleta Jimenez De Aberasturi; Javier Aizpurua; Ramon A. Alvarez-Puebla; Baptiste Auguié; Jeremy J. Baumberg; Guillermo C. Bazan; Steven E. J. Bell; Anja Boisen; Alexandre G. Brolo; et al.Jaebum ChooDana Cialla-MayVolker DeckertLaura FabrisKaren FauldsF. Javier García de AbajoRoyston GoodacreDuncan GrahamAmanda J. HaesChristy L. HaynesChristian HuckTamitake ItohMikael KällJanina KneippNicholas A. KotovHua KuangEric C. Le RuHiang Kwee LeeJian-Feng LiXing Yi LingStefan MaierThomas MayerhoeferMartin MoskovitsKei MurakoshiJwa-Min NamShuming NieYukihiro OzakiIsabel Pastoriza-SantosJorge Perez-JusteJuergen PoppAnnemarie PucciStephanie ReichBin RenGeorge C. SchatzTimur ShegaiSebastian SchlückerLi-Lin TayK. George ThomasZhong-Qun TianRichard P. Van DuyneTuan Vo-DinhYue WangKatherine A. WilletsChuanlai XuHongxing XuYikai XuYuko S. YamamotoBing ZhaoLuis M. Liz-Marzán Present and Future of Surface-Enhanced Raman Scattering. ACS Nano 2019, 14, 28-117, 10.1021/acsnano.9b04224.
- Eric C. Le Ru; Pablo G. Etchegoin; Single-Molecule Surface-Enhanced Raman Spectroscopy. Annual Review of Physical Chemistry 2012, 63, 65-87, 10.1146/annurev-physchem-032511-143757.
- Hae Mi Lee; Seung Min Jin; Hyung Min Kim; Yung Doug Suh; Single-molecule surface-enhanced Raman spectroscopy: a perspective on the current status. Physical Chemistry Chemical Physics 2013, 15, 5276-5287, 10.1039/c3cp44463e.
- Ángela I. López-Lorente; Recent developments on gold nanostructures for surface enhanced Raman spectroscopy: Particle shape, substrates and analytical applications. A review. Analytica Chimica Acta 2021, 1168, 338474, 10.1016/j.aca.2021.338474.
- Wafa Safar; Médéric Lequeux; Jeanne Solard; Alexis P. A. Fischer; Nordin Felidj; Pietro Giuseppe Gucciardi; Mathieu Edely; Marc Lamy De La Chapelle; Gold Nanocylinders on Gold Film as a Multi-spectral SERS Substrate. Nanomaterials 2020, 10, 927, 10.3390/nano10050927.
- Furong Tian; Franck Bonnier; Alan Casey; Anne E. Shanahan; Hugh J. Byrne; Surface enhanced Raman scattering with gold nanoparticles: effect of particle shape. Analytical Methods 2014, 6, 9116-9123, 10.1039/c4ay02112f.
- Qiao Xu; Wei Liu; Li Li; Feng Zhou; Jian Zhou; Yang Tian; Ratiometric SERS imaging and selective biosensing of nitric oxide in live cells based on trisoctahedral gold nanostructures. Chemical Communications 2017, 53, 1880-1883, 10.1039/c6cc09563a.
- Yang Zhang; Ge Wang; Lu Yang; Fei Wang; Aihua Liu; Recent advances in gold nanostructures based biosensing and bioimaging. Coordination Chemistry Reviews 2018, 370, 1-21, 10.1016/j.ccr.2018.05.005.
- Bin Zhao; Jianlei Shen; Shixing Chen; Dongfang Wang; Fan Li; Sanjay Mathur; Shiping Song; Chunhai Fan; Gold nanostructures encoded by non-fluorescent small molecules in polyA-mediated nanogaps as universal SERS nanotags for recognizing various bioactive molecules. Chemical Science 2014, 5, 4460-4466, 10.1039/c4sc01792g.
- M. Fleischmann; P.J. Hendra; A.J. McQuillan; Raman spectra of pyridine adsorbed at a silver electrode. Chemical Physics Letters 1974, 26, 163-166, 10.1016/0009-2614(74)85388-1.
- Huiyuan Guo; Baoshan Xing; Jason C. White; Arnab Mukherjee; Lili He; Ultra-sensitive determination of silver nanoparticles by surface-enhanced Raman spectroscopy (SERS) after hydrophobization-mediated extraction. The Analyst 2016, 141, 5261-5264, 10.1039/c6an01186a.
- Kevin G. Stamplecoskie; Juan C. Scaiano; Vidhu S. Tiwari; Hanan Anis; Optimal Size of Silver Nanoparticles for Surface-Enhanced Raman Spectroscopy. The Journal of Physical Chemistry C 2011, 115, 1403-1409, 10.1021/jp106666t.
- Stacey Laing; Lauren E. Jamieson; Karen Faulds; Duncan Graham; Surface-enhanced Raman spectroscopy for in vivo biosensing. Nature Reviews Chemistry 2017, 1, 0060, 10.1038/s41570-017-0060.
- Mehmet V Yigit; Zdravka Medarova; In vivo and ex vivo applications of gold nanoparticles for biomedical SERS imagingi. American journal of nuclear medicine and molecular imaging 2012, 2, 232-241.
- Vincenzo Amendola; Moreno Meneghetti; What controls the composition and the structure of nanomaterials generated by laser ablation in liquid solution?. Physical Chemistry Chemical Physics 2012, 15, 3027-3046, 10.1039/c2cp42895d.
- Vincenzo Amendola; Moreno Meneghetti; Laser ablation synthesis in solution and size manipulation of noble metal nanoparticles. Physical Chemistry Chemical Physics 2009, 11, 3805-3821, 10.1039/b900654k.
- Mohammad Tahghighi; Davide Janner; Jordi Ignés-Mullol; Optimizing Gold Nanoparticle Size and Shape for the Fabrication of SERS Substrates by Means of the Langmuir–Blodgett Technique. Nanomaterials 2020, 10, 2264, 10.3390/nano10112264.
- Liangbao Yang; Pan Li; Honglin Liu; Xianghu Tang; Jinhuai Liu; A dynamic surface enhanced Raman spectroscopy method for ultra-sensitive detection: from the wet state to the dry state. Chemical Society Reviews 2015, 44, 2837-2848, 10.1039/c4cs00509k.
- Roberto Pilot; Raffaella Signorini; Christian Durante; Laura Orian; Manjari Bhamidipati; Laura Fabris; A Review on Surface-Enhanced Raman Scattering. Biosensors 2019, 9, 57, 10.3390/bios9020057.
- M. Kahl; E. Voges; S. Kostrewa; C. Viets; Wieland Hill; Periodically structured metallic substrates for SERS. Sensors and Actuators B: Chemical 1998, 51, 285-291, 10.1016/s0925-4005(98)00219-6.
- Rokas Šakalys; Kiang Wei Kho; Tia E. Keyes; A reproducible, low cost microfluidic microcavity array SERS platform prepared by soft lithography from a 2 photon 3D printed template. Sensors and Actuators B: Chemical 2021, 340, 129970, 10.1016/j.snb.2021.129970.
- S. N. Terekhov; S. M. Kachan; A. Yu. Panarin; P. Mojzes; Surface-enhanced Raman scattering on silvered porous alumina templates: role of multipolar surface plasmon resonant modes. Physical Chemistry Chemical Physics 2015, 17, 31780-31789, 10.1039/c5cp04197j.
- Jeongwoo Ham; Byung Ju Yun; Won-Gun Koh; SERS-based biosensing platform using shape-coded hydrogel microparticles incorporating silver nanoparticles. Sensors and Actuators B: Chemical 2021, 341, 129989, 10.1016/j.snb.2021.129989.
- Mohammad Tavakkoli Yaraki; Yen Nee Tan; Metal Nanoparticles‐Enhanced Biosensors: Synthesis, Design and Applications in Fluorescence Enhancement and Surface‐enhanced Raman Scattering. Chemistry – An Asian Journal 2020, 15, 3180-3208, 10.1002/asia.202000847.
- Jibin Song; Bo Duan; Chenxu Wang; Jiajing Zhou; Lu Pu; Zheng Fang; Peng Wang; Teik-Thye Lim; Hongwei Duan; SERS-Encoded Nanogapped Plasmonic Nanoparticles: Growth of Metallic Nanoshell by Templating Redox-Active Polymer Brushes. Journal of the American Chemical Society 2014, 136, 6838-6841, 10.1021/ja502024d.
- Vincenzo Amendola; Stefano Scaramuzza; Lucio Litti; Moreno Meneghetti; Gaia Zuccolotto; Antonio Rosato; Elena Nicolato; Pasquina Marzola; Giulio Fracasso; Cristina Anselmi; et al.Marcella PintoMarco Colombatti Magneto-Plasmonic Au-Fe Alloy Nanoparticles Designed for Multimodal SERS-MRI-CT Imaging. Small 2014, 10, 2476-2486, 10.1002/smll.201303372.
- Jingjing Feng; Xiaoling Wu; Wei Ma; Hua Kuang; Liguang Xu; Chuanlai Xu; A SERS active bimetallic core–satellite nanostructure for the ultrasensitive detection of Mucin-1. Chemical Communications 2015, 51, 14761-14763, 10.1039/c5cc05255f.
- Li-An Wu; Wei-En Li; Ding-Zheng Lin; Yih-Fan Chen; Three-Dimensional SERS Substrates Formed with Plasmonic Core-Satellite Nanostructures. Scientific Reports 2017, 7, 1-11, 10.1038/s41598-017-13577-9.
- Michael J. Natan; Concluding Remarks : Surface enhanced Raman scattering. Faraday Discussions 2006, 132, 321-328, 10.1039/b601494c.
- Arun Singh Patel; Subhavna Juneja; Pawan K. Kanaujia; Vikas Maurya; G. Vijaya Prakash; Anirban Chakraborti; Jaydeep Bhattacharya; Gold nanoflowers as efficient hosts for SERS based sensing and bio-imaging. Nano-Structures & Nano-Objects 2018, 16, 329-336, 10.1016/j.nanoso.2018.09.001.
- Shuai He; Jefri Chua; Eddie Khay Ming Tan; James Chen Yong Kah; Optimizing the SERS enhancement of a facile gold nanostar immobilized paper-based SERS substrate. RSC Advances 2017, 7, 16264-16272, 10.1039/c6ra28450g.
- A. S. D. S. Indrasekara; S. Meyers; S. Shubeita; L. C. Feldman; T. Gustafsson; L. Fabris; Gold nanostar substrates for SERS-based chemical sensing in the femtomolar regime. Nanoscale 2014, 6, 8891-8899, 10.1039/c4nr02513j.
- Nahla A. Hatab; C. M. Rouleau; Scott T. Retterer; Gyula Eres; Paul B. Hatzinger; Baohua Gu; An integrated portable Raman sensor with nanofabricated gold bowtie array substrates for energetics detection. The Analyst 2011, 136, 1697-1702, 10.1039/c0an00982b.
- Jian Zhang; Mehrdad Irannejad; Bo Cui; Bowtie Nanoantenna with Single-Digit Nanometer Gap for Surface-Enhanced Raman Scattering (SERS). Plasmonics 2014, 10, 831-837, 10.1007/s11468-014-9870-5.
- Jun Dong; Xing Zhao; Wei Gao; Qingyan Han; Jianxia Qi; Yongkai Wang; Sandong Guo; Mengtao Sun; Nanoscale Vertical Arrays of Gold Nanorods by Self-Assembly: Physical Mechanism and Application. Nanoscale Research Letters 2019, 14, 118, 10.1186/s11671-019-2946-6.
- Kanyawan Ponlamuangdee; Gabor L. Hornyak; Tanujjal Bora; Suwussa Bamrungsap; Graphene oxide/gold nanorod plasmonic paper – a simple and cost-effective SERS substrate for anticancer drug analysis. New J. Chem. 2020, 44, 14087-14094, 10.1039/d0nj02448a.
- Xiangxian Wang; Xuelin Bai; Zhiyuan Pang; Jiankai Zhu; Yuan Wu; Hua Yang; Yunping Qi; Xiaolei Wen; Surface-enhanced Raman scattering by composite structure of gold nanocube-PMMA-gold film. Optical Materials Express 2019, 9, 1872-1881, 10.1364/ome.9.001872.
- Shuang Lin; Xiang Lin; Yuxin Shang; Siqingaowa Han; Wuliji Hasi; Li Wang; Self-Assembly of Faceted Gold Nanocrystals for Surface-Enhanced Raman Scattering Application. The Journal of Physical Chemistry C 2019, 123, 24714-24722, 10.1021/acs.jpcc.9b06686.
- Daedu Lee; Sangwoon Yoon; Gold Nanocube–Nanosphere Dimers: Preparation, Plasmon Coupling, and Surface-Enhanced Raman Scattering. The Journal of Physical Chemistry C 2015, 119, 7873-7882, 10.1021/acs.jpcc.5b00314.
- Virginia Joseph; Andrea Matschulat; Jörg Polte; Simone Rolf; Franziska Emmerling; Janina Kneipp; SERS enhancement of gold nanospheres of defined size. Journal of Raman Spectroscopy 2011, 42, 1736-1742, 10.1002/jrs.2939.
- Wonjoo Lee; Seung Yong Lee; Robert M. Briber; Oded Rabin; Self-Assembled SERS Substrates with Tunable Surface Plasmon Resonances. Advanced Functional Materials 2011, 21, 3424-3429, 10.1002/adfm.201101218.
- N. Félidj; J. Aubard; G. Lévi; Joachim Krenn; Marco Salerno; G. Schider; B. Lamprecht; A. Leitner; F. R. Aussenegg; Controlling the optical response of regular arrays of gold particles for surface-enhanced Raman scattering. Physical Review B 2002, 65, 075419, 10.1103/physrevb.65.075419.
- Mohammad Kamal Hossain; Nanoassembly of gold nanoparticles: An active substrate for size-dependent surface-enhanced Raman scattering. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy 2020, 242, 118759, 10.1016/j.saa.2020.118759.
- V. Weber; A. Feis; C. Gellini; R. Pilot; P. R. Salvi; R. Signorini; Far- and near-field properties of gold nanoshells studied by photoacoustic and surface-enhanced Raman spectroscopies. Physical Chemistry Chemical Physics 2014, 17, 21190-21197, 10.1039/c4cp05054a.
- Sehee Jeong; Min-Woo Kim; Yong-Ryun Jo; Na-Yeong Kim; Dooho Kang; Seong Youl Lee; Sang-Youp Yim; Bong-Joong Kim; Joon Heon Kim; Hollow Porous Gold Nanoshells with Controlled Nanojunctions for Highly Tunable Plasmon Resonances and Intense Field Enhancements for Surface-Enhanced Raman Scattering. ACS Applied Materials & Interfaces 2019, 11, 44458-44465, 10.1021/acsami.9b16983.
- Tae-Hyeon Park; Du-Jeon Jang; Laser-induced fabrication of porous gold nanoshells. Nanoscale 2018, 10, 20108-20112, 10.1039/c8nr04617d.
- R. V. William; G. M. Das; V. R. Dantham; R. Laha; Enhancement of Single Molecule Raman Scattering using Sprouted Potato Shaped Bimetallic Nanoparticles. Scientific Reports 2019, 9, 1-12, 10.1038/s41598-019-47179-4.
- Katherine C. Grabar; R. Griffith. Freeman; Michael B. Hommer; Michael J. Natan; Preparation and Characterization of Au Colloid Monolayers. Analytical Chemistry 1995, 67, 735-743, 10.1021/ac00100a008.
- R. Griffith Freeman; Katherine C. Grabar; Keith J. Allison; Robin M. Bright; Jennifer A. Davis; Andrea P. Guthrie; Michael B. Hommer; Michael A. Jackson; Patrick C. Smith; Daniel G. Walter; et al.Michael J. Natan Self-Assembled Metal Colloid Monolayers: An Approach to SERS Substrates. Science 1995, 267, 1629-1632, 10.1126/science.267.5204.1629.
- Barbara Fazio; Cristiano D'Andrea; Antonino Foti; Elena Messina; Alessia Irrera; Maria Grazia Donato; Valentina Villari; Norberto Micali; Onofrio M. Marago; Pietro G. Gucciardi; et al. SERS detection of Biomolecules at Physiological pH via aggregation of Gold Nanorods mediated by Optical Forces and Plasmonic Heating. Scientific Reports 2016, 6, 26952, 10.1038/srep26952.
- Antonino Foti; Cristiano D’Andrea; Valentina Villari; Norberto Micali; Maria Grazia Donato; Barbara Fazio; Onofrio M. Maragò; Raymond Gillibert; Marc Lamy De La Chapelle; Pietro G. Gucciardi; et al. Optical Aggregation of Gold Nanoparticles for SERS Detection of Proteins and Toxins in Liquid Environment: Towards Ultrasensitive and Selective Detection. Materials 2018, 11, 440, 10.3390/ma11030440.
- Anni Lehmuskero; Peter Johansson; Halina Rubinsztein-Dunlop; Lianming Tong; Mikael Käll; Laser Trapping of Colloidal Metal Nanoparticles. ACS Nano 2015, 9, 3453-3469, 10.1021/acsnano.5b00286.
- Hyungsoon Im; Kyle C. Bantz; Nathan C. Lindquist; Christy Haynes; Sang-Hyun Oh; Vertically Oriented Sub-10-nm Plasmonic Nanogap Arrays. Nano Letters 2010, 10, 2231-2236, 10.1021/nl1012085.
- Lisheng Zhang; Fujun Xu; Jiaming Wang; Chenguang He; Weiwei Guo; Mingxing Wang; Bowen Sheng; Lin Lu; Zhixin Qin; Xinqiang Wang; et al.Bo Shen High-quality AlN epitaxy on nano-patterned sapphire substrates prepared by nano-imprint lithography. Scientific Reports 2016, 6, 35934, 10.1038/srep35934.
- Vignesh Suresh; Lu Ding; Ah Bian Chew; Fung Ling Yap; Fabrication of Large-Area Flexible SERS Substrates by Nanoimprint Lithography. ACS Applied Nano Materials 2018, 1, 886-893, 10.1021/acsanm.7b00295.
- Nestor Gisbert Quilis; Médéric Lequeux; Priyamvada Venugopalan; Imran Khan; Wolfgang Knoll; Souhir Boujday; Marc Lamy de la Chapelle; Jakub Dostalek; Tunable laser interference lithography preparation of plasmonic nanoparticle arrays tailored for SERS. Nanoscale 2018, 10, 10268-10276, 10.1039/c7nr08905h.
- George M. Whitesides; Bartosz Grzybowski; Self-Assembly at All Scales. Science 2002, 295, 2418-2421, 10.1126/science.1070821.
- Marco Pisco; Francesco Galeotti; Giuseppe Quero; Giorgio Grisci; Alberto Micco; Lucia V Mercaldo; Paola Delli Veneri; Antonello Cutolo; Andrea Cusano; Nanosphere lithography for optical fiber tip nanoprobes. Light: Science & Applications 2016, 6, e16229-e16229, 10.1038/lsa.2016.229.
- Chengpeng Zhang; Peiyun Yi; Linfa Peng; Xinmin Lai; Jie Chen; Meizhen Huang; Jun Ni; Continuous fabrication of nanostructure arrays for flexible surface enhanced Raman scattering substrate. Scientific Reports 2017, 7, 39814, 10.1038/srep39814.
- Giuseppe Quero; Gianluigi Zito; Stefano Managò; Francesco Galeotti; Marco Pisco; Anna Chiara De Luca; Andrea Cusano; Nanosphere Lithography on Fiber: Towards Engineered Lab-On-Fiber SERS Optrodes. Sensors 2018, 18, 680, 10.3390/s18030680.
- Ian Bruzas; William Lum; Zohre Gorunmez; Laura Sagle; Advances in surface-enhanced Raman spectroscopy (SERS) substrates for lipid and protein characterization: sensing and beyond. The Analyst 2018, 143, 3990-4008, 10.1039/c8an00606g.
- Kevin M. Cheung; John M. Abendroth; Nako Nakatsuka; Bowen Zhu; Yang Yang; Anne M. Andrews; Paul S. Weiss; Detecting DNA and RNA and Differentiating Single-Nucleotide Variations via Field-Effect Transistors. Nano Letters 2020, 20, 5982-5990, 10.1021/acs.nanolett.0c01971.
- Tahminur Rahman; Muhammed Salah Uddin; Razia Sultana; Arumina Moue; Muntahina Setu; Polymerase Chain Reaction (PCR): A Short Review. Anwer Khan Modern Medical College Journal 2013, 4, 30-36, 10.3329/akmmcj.v4i1.13682.
- Jin-Mei Yang; Lei Jin; Zhong-Qin Pan; Yue Zhou; Hai-Ling Liu; Li-Na Ji; Xing-Hua Xia; Kang Wang; Surface-Enhanced Raman Scattering Probing the Translocation of DNA and Amino Acid through Plasmonic Nanopores. Analytical Chemistry 2019, 91, 6275-6280, 10.1021/acs.analchem.9b01045.
- Woo Hyun Kim; Jong Uk Lee; Sojin Song; Soohyun Kim; Young Jae Choi; Sang Jun Sim; A label-free, ultra-highly sensitive and multiplexed SERS nanoplasmonic biosensor for miRNA detection using a head-flocked gold nanopillar. The Analyst 2019, 144, 1768-1776, 10.1039/c8an01745j.
- Danny van Lierop; Karen Faulds; Duncan Graham; Separation Free DNA Detection Using Surface Enhanced Raman Scattering. Analytical Chemistry 2011, 83, 5817-5821, 10.1021/ac200514e.
- Xinpan Wei; Shao Su; Yuanyuan Guo; Xiangxu Jiang; Yiling Zhong; Yuanyuan Su; Chunhai Fan; Shuit-Tong Lee; Yao He; A Molecular Beacon-Based Signal-Off Surface-Enhanced Raman Scattering Strategy for Highly Sensitive, Reproducible, and Multiplexed DNA Detection. Small 2013, 9, 2493-2499, 10.1002/smll.201202914.
- Kholud Dardir; Hao Wang; Brigitte E. Martin; Maria Atzampou; Christopher B. Brooke; Laura Fabris; SERS Nanoprobe for Intracellular Monitoring of Viral Mutations. The Journal of Physical Chemistry C 2020, 124, 3211-3217, 10.1021/acs.jpcc.9b09253.
- Sujuan Ye; Menglei Wang; Zhenxing Wang; Na Zhang; Xiliang Luo; A DNA–linker–DNA bifunctional probe for simultaneous SERS detection of miRNAs via symmetric signal amplification. Chemical Communications 2018, 54, 7786-7789, 10.1039/c8cc02910e.
- Lamyaa M. Almehmadi; Stephanie M. Curley; Natalya A. Tokranova; Scott A. Tenenbaum; Igor K. Lednev; Surface Enhanced Raman Spectroscopy for Single Molecule Protein Detection. Scientific Reports 2019, 9, 1-9, 10.1038/s41598-019-48650-y.
- Thibault Brulé; Alexandre Bouhelier; Alain Dereux; Eric Finot; Discrimination between Single Protein Conformations Using Dynamic SERS. ACS Sensors 2016, 1, 676-680, 10.1021/acssensors.6b00097.
- Adem Zengin; Ugur Tamer; Tuncer Caykara; A SERS-Based Sandwich Assay for Ultrasensitive and Selective Detection of Alzheimer’s Tau Protein. Biomacromolecules 2013, 14, 3001-3009, 10.1021/bm400968x.
- Kazuki Bando; Nicholas Smith; Jun Ando; Katsumasa Fujita; Satoshi Kawata; Analysis of dynamic SERS spectra measured with a nanoparticle during intracellular transportation in 3D. Journal of Optics 2015, 17, 114023, 10.1088/2040-8978/17/11/114023.
- Qian Liu; Jiahong Wang; Beike Wang; Zhe Li; Hao Huang; Chengzhang Li; Xuefeng Yu; Paul K. Chu; Paper-based plasmonic platform for sensitive, noninvasive, and rapid cancer screening. Biosensors and Bioelectronics 2013, 54, 128-134, 10.1016/j.bios.2013.10.067.
- Gustavo Bodelon; Verónica Montes García; Cristina Fernández-López; Isabel Pastoriza-Santos; Jorge Pérez-Juste; Luis M. Liz-Marzán; Au@pNIPAM SERRS Tags for Multiplex Immunophenotyping Cellular Receptors and Imaging Tumor Cells. Small 2015, 11, 4149-4157, 10.1002/smll.201500269.
- Marjorie R. Willner; Kay S. McMillan; Duncan Graham; Peter J. Vikesland; Michele Zagnoni; Surface-Enhanced Raman Scattering Based Microfluidics for Single-Cell Analysis. Analytical Chemistry 2018, 90, 12004-12010, 10.1021/acs.analchem.8b02636.
- Javier Plou; Isabel García; Mathias Charconnet; Ianire Astobiza; Clara García-Astrain; Cristiano Matricardi; Agustin Mihi; Arkaitz Carracedo; Luis M. Liz‐Marzán; Multiplex SERS Detection of Metabolic Alterations in Tumor Extracellular Media. Advanced Functional Materials 2020, 30, 1910335, 10.1002/adfm.201910335.
- Stefano Managò; Gianluigi Zito; Alessandra Rogato; Maurizio Casalino; Emanuela Esposito; Anna Chiara De Luca; Edoardo De Tommasi; Bioderived Three-Dimensional Hierarchical Nanostructures as Efficient Surface-Enhanced Raman Scattering Substrates for Cell Membrane Probing. ACS Applied Materials & Interfaces 2018, 10, 12406-12416, 10.1021/acsami.7b19285.
- Anna Chiara De Luca; Peter Reader-Harris; Michael Mazilu; Stefania Mariggiò; Daniela Corda; Andrea Di Falco; Reproducible Surface-Enhanced Raman Quantification of Biomarkers in Multicomponent Mixtures. ACS Nano 2014, 8, 2575-2583, 10.1021/nn406200y.
- Ya-Fei Tian; Cui-Fang Ning; Fang He; Bin-Cheng Yin; Bang-Ce Ye; Highly sensitive detection of exosomes by SERS using gold nanostar@Raman reporter@nanoshell structures modified with a bivalent cholesterol-labeled DNA anchor. The Analyst 2018, 143, 4915-4922, 10.1039/c8an01041b.
- Stefano Managò; Chiara Tramontano; Donatella Delle Cave; Giovanna Chianese; Gianluigi Zito; Luca De Stefano; Monica Terracciano; Enza Lonardo; Anna Chiara De Luca; Ilaria Rea; et al. SERS Quantification of Galunisertib Delivery in Colorectal Cancer Cells by Plasmonic‐Assisted Diatomite Nanoparticles. Small 2021, 17, 2101711, 10.1002/smll.202101711.
- Xiaoxia Wu; Yuanzhi Xia; Youju Huang; Juan Li; Huimin Ruan; Tianxiang Chen; Liqiang Luo; Zheyu Shen; Aiguo Wu; Improved SERS-Active Nanoparticles with Various Shapes for CTC Detection without Enrichment Process with Supersensitivity and High Specificity. ACS Applied Materials & Interfaces 2016, 8, 19928-19938, 10.1021/acsami.6b07205.
More
Information
Subjects:
Spectroscopy; Nanoscience & Nanotechnology; Optics
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.0K
Revisions:
2 times
(View History)
Update Date:
01 Nov 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




