Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Everett J. Moding | + 772 word(s) | 772 | 2021-10-26 10:28:38 |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Moding, E. Molecular Radiobiology with Liquid Biopsies. Encyclopedia. Available online: https://encyclopedia.pub/entry/15521 (accessed on 08 February 2026).
Moding E. Molecular Radiobiology with Liquid Biopsies. Encyclopedia. Available at: https://encyclopedia.pub/entry/15521. Accessed February 08, 2026.
Moding, Everett. "Molecular Radiobiology with Liquid Biopsies" Encyclopedia, https://encyclopedia.pub/entry/15521 (accessed February 08, 2026).
Moding, E. (2021, October 29). Molecular Radiobiology with Liquid Biopsies. In Encyclopedia. https://encyclopedia.pub/entry/15521
Moding, Everett. "Molecular Radiobiology with Liquid Biopsies." Encyclopedia. Web. 29 October, 2021.
Copy Citation
Using liquid biopsy techniques including ctDNA analysis as tools to study radiobiology in patients with cancer.
liquid biopsies
radiation biology
tumor evolution
1. Unique Insights into Tumor Heterogeneity and Clonal Evolution
In addition to being a powerful biomarker to enable personalized cancer therapy, liquid biopsies provide a unique window into tumor biology that can be sampled at multiple time points over the course of the natural history of disease and throughout therapy. Recent studies have highlighted the substantial genetic heterogeneity that can exist with human tumors[1]. The accumulation of mutations, genetic drift, and selective pressures within the tumor microenvironment can lead to tumor evolution that can impact how cancers respond to treatment[2]. Multi-region sequencing has suggested that early tumor development may generally occur under weak evolutionary pressures[3][4] but that selection appears to increase at later stages of tumor development and metastasis[5][6][7]. However, the impact of intratumoral heterogeneity on radiotherapy outcomes remains to be defined. Modeling experiments suggest that subclones with heterogenous radiosensitivity could be a major determinant in treatment response[8]. Further, although there is strong evidence that systemic therapy selects resistant subclones that lead to disease progression[9], the effect of radiotherapy on tumor evolution in acquired resistance to radiotherapy is less clear. The sequencing of pre- and post-treatment samples from patients undergoing chemoradiotherapy for rectal cancer suggested that radiotherapy can substantially impact the clonal architecture and can potentially select resistant subclones[10]. Additionally, retrospective clinical studies have reported worse local control after reirradiation compared to initial treatment with radiotherapy[11][12], but these studies are confounded by lower doses of radiation in the reirradiation setting due to concern for toxicity.
Two major limitations to studying clonal evolution during radiotherapy are (1) sampling biases related to analyzing one or a few regions within a patient’s tumor and (2) a lack of longitudinal specimens to study clonal dynamics. ctDNA has the potential to be released from all of the tumor cells in a patient and can be sampled longitudinally from routine blood draws, making it a powerful tool to characterize clonal evolution (Figure 1). Multiple studies have applied ctDNA analysis to monitor clonal evolution during the treatment of patients with systemic therapy. For example, analysis of paired pre-treatment and end-of-treatment ctDNA samples demonstrated clear evidence of resistant subclone selection during the treatment of metastatic estrogen receptor-positive breast cancer with CDK4/6 inhibition and endocrine therapy[13]. In addition, ctDNA analysis demonstrated that subclonal tumor populations contribute to relapse after surgery and adjuvant chemotherapy for early stage lung cancer[14]. As a result, longitudinal ctDNA analysis and subclone tracking could help to clarify the modes of tumor evolution during radiotherapy.
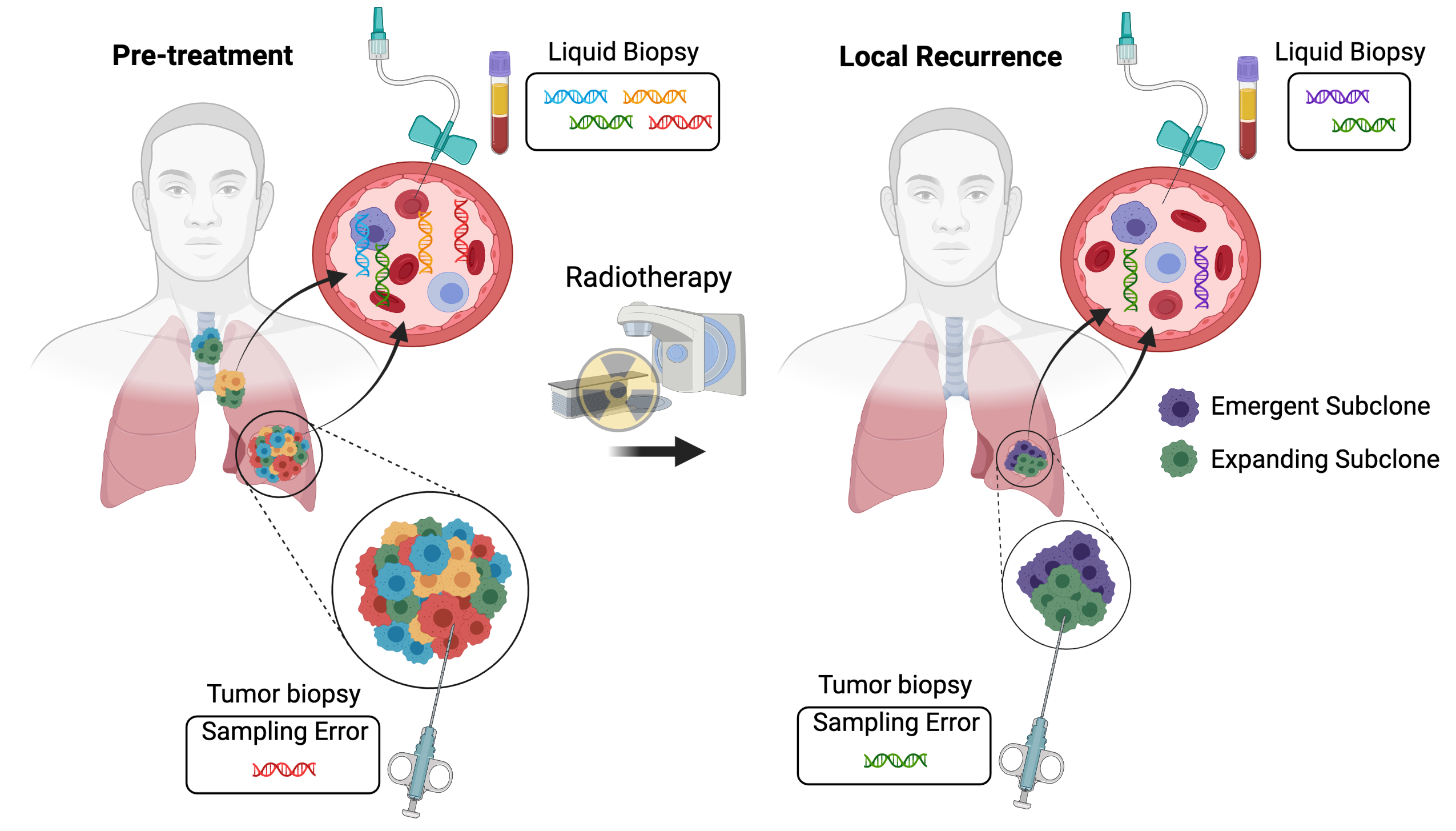
Figure 1. Interrogating tumor heterogeneity, clonal evolution, and resistance to radiotherapy using liquid biopsies. Tumors consist of multiple subclones with distinct genetic alterations. Tumor biopsies sample a single region of one tumor deposit. As a result, resistant subclones may be missed due to sampling error. In contrast, liquid biopsies may characterize the full extent of intratumoral heterogeneity by sampling circulating tumor DNA released from all of the tumor deposits within a patient. Liquid biopsies can also be repeated at multiple time points, enabling the longitudinal tracking of clonal evolution during radiotherapy and the identification of emergent and expanding tumor subclones that mediate radiation resistance. Created with BioRender.com.
2. Identifying Mediators of Radiotherapy Resistance and Sensitivity
Innovative approaches will be necessary to identify novel mediators of radiotherapy resistance that can inform precision therapy. In most cancers, the first assessment of treatment response after radiotherapy does not occur until months after the completion of treatment, making it challenging to identify rapid responders. The radiation-induced inflammatory and fibrotic tissue changes described above also complicate early response assessment to radiotherapy with imaging. ctDNA kinetics during treatment represent novel markers of treatment response that may enable the identification of rapid responders[15]. Characterization of these ctDNA rapid responders could help to identify the molecular signatures that underly radiosensitivity to enable the pre-treatment identification of patients who will respond favorably to treatment.
In addition to elucidating the biology of upfront treatment response, liquid biopsies can help to identify expanding and emergent mutations that may contribute to treatment resistance. For example, a prospective study of patients undergoing targeted therapy for gastrointestinal cancers found that ctDNA analysis identified resistance alterations not identified from matched tumor sequencing in 78% of cases[16]. Furthermore, multiple resistance alterations were identified in 40% of patients by ctDNA analysis compared to only 9% of patients by matched tumor sequencing. Several additional studies have also leveraged ctDNA analysis to identify mechanisms of resistance to systemic therapy[17][18][19][20]. Taken together, the liquid biopsy approaches discussed in this entry improve upon existing methodologies for studying tumor evolution and provide a novel opportunity to investigate tumor radiation biology.
References
- McGranahan, N.; Swanton, C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 2015, 27, 15–26.
- Turajlic, S.; Sottoriva, A.; Graham, T.; Swanton, C. Resolving genetic heterogeneity in cancer. Nat. Rev. Genet. 2019, 20, 404–416.
- Sottoriva, A.; Kang, H.; Ma, Z.; Graham, T.A.; Salomon, M.P.; Zhao, J.; Marjoram, P.; Siegmund, K.; Press, M.F.; Shibata, D.; et al. A Big Bang model of human colorectal tumor growth. Nat. Genet. 2015, 47, 209–216.
- Ling, S.; Hu, Z.; Yang, Z.; Yang, F.; Li, Y.; Lin, P.; Chen, K.; Dong, L.; Cao, L.; Tao, Y.; et al. Extremely high genetic diversity in a single tumor points to prevalence of non-Darwinian cell evolution. Proc. Natl. Acad. Sci. USA 2015, 112, E6496–E6505.
- Reiter, J.G.; Hung, W.-T.; Lee, I.-H.; Nagpal, S.; Giunta, P.; Degner, S.; Liu, G.; Wassenaar, E.C.E.; Jeck, W.R.; Taylor, M.S.; et al. Lymph node metastases develop through a wider evolutionary bottleneck than distant metastases. Nat. Genet. 2020, 52, 692–700.
- Reiter, J.G.; Makohon-Moore, A.P.; Gerold, J.M.; Heyde, A.; Attiyeh, M.A.; Kohutek, Z.A.; Tokheim, C.J.; Brown, A.; DeBlasio, R.M.; Niyazov, J.; et al. Minimal functional driver gene heterogeneity among untreated metastases. Science 2018, 361, 1033–1037.
- Sun, R.; Hu, Z.; Sottoriva, A.; Graham, T.A.; Harpak, A.; Ma, Z.; Fischer, J.M.; Shibata, D.; Curtis, C. Between-region genetic divergence reflects the mode and tempo of tumor evolution. Nat. Genet. 2017, 49, 1015–1024.
- Alfonso, J.C.L.; Berk, L. Modeling the effect of intratumoral heterogeneity of radiosensitivity on tumor response over the course of fractionated radiation therapy. Radiat. Oncol. 2019, 14, 88.
- Greaves, M.; Maley, C.C. Clonal evolution in cancer. Nature 2012, 481, 306–313.
- Frydrych, L.M.; Ulintz, P.; Bankhead, A.; Sifuentes, C.; Greenson, J.; Maguire, L.; Irwin, R.; Fearon, E.R.; Hardiman, K.M. Rectal cancer sub-clones respond differentially to neoadjuvant therapy. Neoplasia 2019, 21, 1051–1062.
- Indelicato, D.J.; Meadows, K.; Gibbs, C.P.; Morris, C.G.; Scarborough, M.T.; Zlotecki, R.A. Effectiveness and morbidity associated with reirradiation in conservative salvage management of recurrent soft-tissue sarcoma. Int. J. Radiat. Oncol. Biol. Phys. 2009, 73, 267–272.
- Torres, M.A.; Ballo, M.T.; Butler, C.E.; Feig, B.W.; Cormier, J.N.; Lewis, V.O.; Pollock, R.E.; Pisters, P.W.; Zagars, G.K. Management of locally recurrent soft-tissue sarcoma after prior surgery and radiation therapy. Int. J. Radiat. Oncol. Biol. Phys. 2007, 67, 1124–1129.
- O’Leary, B.; Cutts, R.J.; Liu, Y.; Hrebien, S.; Huang, X.; Fenwick, K.; André, F.; Loibl, S.; Loi, S.; Garcia-Murillas, I.; et al. The genetic landscape and clonal evolution of breast cancer resistance to palbociclib plus fulvestrant in the PALOMA-3 trial. Cancer Discov. 2018, 8, 1390–1403.
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446–451.
- Moding, E.J.; Liu, Y.; Hui, A.B.; He, J.; Qiao, Y.; Xu, T.; Yao, L.; Gandhi, S.; Liao, Z.; Das, M.; et al. Abstract PO-069: Circulating tumor DNA kinetics to identify genomic predictors of rapid response to chemoradiation in non-small cell lung cancer. Clin. Cancer Res. 2021, 27, PO-069.
- Parikh, A.R.; Leshchiner, I.; Elagina, L.; Goyal, L.; Levovitz, C.; Siravegna, G.; Livitz, D.; Rhrissorrakrai, K.; Martin, E.E.; Van Seventer, E.E.; et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 2019, 25, 1415–1421.
- Chabon, J.J.; Simmons, A.D.; Lovejoy, A.F.; Esfahani, M.S.; Newman, A.M.; Haringsma, H.J.; Kurtz, D.M.; Stehr, H.; Scherer, F.; Karlovich, C.A.; et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat. Commun. 2016, 7, 11815.
- Murtaza, M.; Dawson, S.-J.; Tsui, D.W.Y.; Gale, D.; Forshew, T.; Piskorz, A.M.; Parkinson, C.; Chin, S.-F.; Kingsbury, Z.; Wong, A.S.C.; et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013, 497, 108.
- O’Leary, B.; Hrebien, S.; Morden, J.P.; Beaney, M.; Fribbens, C.; Huang, X.; Liu, Y.; Bartlett, C.H.; Koehler, M.; Cristofanilli, M.; et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat. Commun. 2018, 9, 896.
- Khan, K.H.; Cunningham, D.; Werner, B.; Vlachogiannis, G.; Spiteri, I.; Heide, T.; Mateos, J.F.; Vatsiou, A.; Lampis, A.; Damavandi, M.D.; et al. Longitudinal liquid biopsy and mathematical modeling of clonal evolution forecast time to treatment failure in the PROSPECT-C phase II colorectal cancer clinical trial. Cancer Discov. 2018, 8, 1270–1285.
More
Information
Subjects:
Oncology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
878
Revision:
1 time
(View History)
Update Date:
29 Oct 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




