Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ahsan Irshad | + 2756 word(s) | 2756 | 2021-10-09 08:52:01 | | | |
| 2 | Camila Xu | Meta information modification | 2756 | 2021-10-27 04:32:39 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Irshad, A. Soluble Starch Synthase Enzymes in Cereals. Encyclopedia. Available online: https://encyclopedia.pub/entry/15444 (accessed on 07 February 2026).
Irshad A. Soluble Starch Synthase Enzymes in Cereals. Encyclopedia. Available at: https://encyclopedia.pub/entry/15444. Accessed February 07, 2026.
Irshad, Ahsan. "Soluble Starch Synthase Enzymes in Cereals" Encyclopedia, https://encyclopedia.pub/entry/15444 (accessed February 07, 2026).
Irshad, A. (2021, October 27). Soluble Starch Synthase Enzymes in Cereals. In Encyclopedia. https://encyclopedia.pub/entry/15444
Irshad, Ahsan. "Soluble Starch Synthase Enzymes in Cereals." Encyclopedia. Web. 27 October, 2021.
Copy Citation
Starch is synthesized by different enzymes, but starch structure and amount are mainly determined by the activities of starch synthase enzymes (SS) with the involvement of starch branching enzymes (SBEs) and debranching enzymes (DBEs).
amylopectin
cereals
starch synthase
phosphorylation
1. Introduction
Starch is the primary source of energy for human nutrition and is a main product of plant photosynthetic C fixation [1]. Higher plants synthesize storage starch in the form of granules and store in the seeds and tubers. Starch present in these organs and accumulate during the developments of these organs and its stable for long period of time in dry condition. Most of the starch in seeds store in the endosperm tissue with little amount of starch store in embryo and pericarp. Transitory starch present in leaves of plants and is derived from surplus sugar produced during photosynthesis [2]. Natural sugar which is actually a glucose, development in plants is due to degradation of transitory starch which is transported into the cytosol. Starch plays an essential role in plant physiology and alteration of starch levels affect plant growth, seed yield, and flowering time [3]. The degradation of starch occurs during respiration in plants and contributes to the formation of sucrose. This sucrose is transported to the rest of the plant to provide energy in plant growth [4].
Starch is the major polysaccharide in plants, and is composed of two glucan polymers, amylose, and amylopectin. Amylose is a smaller polymer of α-1,4-linked glucose. While amylopectin is highly branched molecule and major component with α-1,4-linked glucose linear chains and α-1,6-linked branched points. The contribution of amylopectin in starch granule is 75% [5]. Starch is formed from the activated nucleotide diphosphate sugar precursor adenosine-5′-diphosphoglucose (ADP-Glc). ADP-Glc is used for elongation of glucan chains by soluble starch synthase (SS) and non-soluble granule bound SS (GBSS) in amylopectin and amylose synthesis, respectively. These α-1,4-linked glucan chains are branched by the introduction of α-1,6-linked branch points with the coordination of starch branching enzymes (SBE). By trimming at specific points in the nascent granules through starch debranching enzymes (DBE), crystalline starch granules are produced. It is accepted that amylopectin branching frequency and pattern is non-random. These glucan chains are categorized with in each molecule on the basis of their connection to other glucan chains: the external chains that have no branches themselves are A-chains. Similarly, B-chains have one or more clusters (B1, B2, B3). The C chain is the part of B-chain in a molecule with free reducing end. The frequency distribution of chain length shows that mostly chains consist of 20–30 glucose units and these are A- and B1-chains in the cluster of amylopectin [6] (Figure 1). Similarly, there are protein targeting to starch (PTST) enzymes (PTST2 and PTST3) which take part in granule initiation in plants and loss of these enzymes causes reduced number of granules in chloroplast. In plants, SSs are GT-B-fold glycosyltransferases, classified within family GT5 in the CAZy database. The archaeal and bacterial GS are the closest counterparts of plant SSs in the GT5 family [7], implying that this family is ancient. All of them use ADP-glucose as nucleotide donor sugar. However, GS in other eukaryotes, such as fungi, yeast and animals, are distantly related to plant SSs, and belong to the GT3 family in the CAZy classification, using UDP-glucose as donor [8].
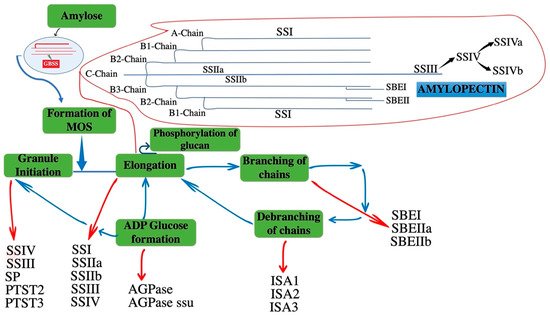
Figure 1. A schematic of enzyme-mediated reactions involved in the formation of starch, amylose, and amylopectin. The diagram represents the interconnection of non-linear reactions of different enzymes during starch biosynthesis. Each class is highlighted with an arrow that is showing each stage of starch biosynthesis with different enzymes. The red arrows mentioned the enzymes of different stages and blue arrows mentioned the relation of different stages during starch formation. ADPglucose pyrophosphorylase: AGPase; ADPglucose pyrophosphorylase small subunit: AGPase ssu; Isoamylase-type debranching enzyme 1, 2, 3: ISA1, ISA2, ISA3; Starch synthase I, IIa, IIb, III, IV: SSI, SSIIa, SSIIb, SSIII, SSIV; Protein targeting to starch: PTST2, PTST2; Starch phosphorylase: SP; Starch branching enzyme I, IIa, IIb: SBEI, SBEIIa, SBEIIb; Granule bound starch synthase: GBSS.
Most of the enzyme classes described have multiple isoforms with overlapping functions [9]. Soluble SSs (SSI, SSII, SSIII, and SSIV) function in the process of starch synthesis have been elucidated by mutant analysis of monocots by using cereal models and of dicots through studying potato tubers, Arabidopsis leaves, and pea embryos. In Arabidopsis, it regulates the granules numbers that form in the chloroplast and it is closely related to SSIV. SSV is a noncanonical isoform with no catalytic glycotransferase activity [10]. The structure and size of amylopectin clusters are mainly controlled by three soluble SSs (SSI, SSII, and SSIII), with the interconnection of SBE and DBE enzymes (Figure 1) [11]. Many SSs genes are present in cereal crops, and their copy number is different in each cereal, presumably reflecting gene duplication, deletion, and genomic polyploidization during evolution (Table 1).
Table 1. Starch synthase genes in cereal crops.
| Species | No. of SS Genes | Gene Names with Accession No./ID | Reference |
|---|---|---|---|
| Hordeum vulgare | 6 | GBSSI (AAM560327.2), SSI (AAF37876), SSII (AAN28307), SSIIIa (AAF87999), SSIIIb (AAL40942), SSIV (AAK97773) | [12] |
| Oryza sativa | 11 | GBSSI (AB425323), GBSSII (AY069940), SSI (AY299404), SSIIa (AF419099), SSIIb (AF395537), SSIIc (AF383878), SSIIIa (AY100469), SSIIIb (AF432915), SSIVa (AY373257), SSIVb (AY373258), SSV (EU621837.1) | [13] |
| Sorghum bicolor | 10 | GBSSI (LOC8068390), GBSSII, SSI (NC054143), SSIIa (EU620718), SSIIb (EU620719), SSIIIa (EU620720), SSIIIb (EU620721), SSIV, SSV (HQ661801) | [14] |
| Triticum aestivum | 7 | GBSSI (AF286320), GBSSII (AF109395), SSI (AJ269503), SSII (AJ269503), SSIIIa (AF258608), SSIIIb (EU333946), SSIV (AY044844) | [15] |
| Zea mays | 10 | GBSSI (AY109531), GBSSII (EF471312), SSI (AF036891), SSIIa (AF019296), SSIIb (EF472249), SSIIc (EU284113), SSIIIa (AF023159), SSIIIb (EF472250), SSIV (EU599036), SSV (NM_001 130131.1) | [16] |
2. Mode of Action and Properties of Soluble SSs in Amylopectin Formation
For the elongation of the α-glucan chain during amylopectin synthesis, three enzymes (SSI, SSII, and SSIII) play important role (Figure 1). Similarly, SSIV is involved in granule initiation and shows close relation to SSIII [17]. SSI, SSII, and SSIII elongate α-glucan chains during amylopectin synthesis with increasingly higher DP (degree of polymerization). SSI synthesizes α-glucan chains from short to intermediate sizes of DP8-12, which are then used as the substrate of SSII to manufacture longer chains of DP12–30. Similarly, SSIII produces long chains of DP ≥ 30 [18]. The products and substrate of these SS isoforms are generalized, and it is inferred from the data of the mutant studies in monocots, dicots, and also upon in vitro biochemical analysis [19]. These three isoforms play an essential role in defining the structure of amylopectin by cooperating with SBEs and DBEs [11] (Figure 1). Although there are variations in glucan chains of different species, the glucan chains found in amylopectin clusters are characteristically of short to medium length appropriate for SSII activity [20]. The binding ability of SSI increases dramatically with the length of substrate chains and is inversely proportional to the catalytic capability of an enzyme. SSIII is thought to be involved in connecting amylopectin clusters because organisms lacking SSIII showed a significant reduction in length of cluster-spanning B chains (B2–3) [21].
Tissue-specific isoforms of SSII and SSIII are present in cereals. These isoforms are thought to be involved in long- or short-chain starch synthesis in different heterotrophic and autotrophic cell types (Table 1) [22]. It is believed that structural variation between starches from different sources is due to the relative contribution of each SS class in various tissues and among species [23]. Due to action of multiple enzymes and alteration of biosynthetic pathways help to cause these structural variations.
2.1. SS Mutants Vital Roles in the Formation Amylopectin Chains: Starch Synthase SSI to SSIII
Loss of SSI activity causes distinct variation in chain length distribution of amylopectin, particularly in A- and B1 chains that help to construct amylopectin clusters. Amylopectin from the endosperm of a ssI rice mutant have shorter chains with DPs of 6–7, while there were fewer chains of DP 8-12 [24]. Similar results were observed in Arabidopsis mutant ssI [25]. In maize, rice, and wheat, ssI mutants possessed short chains with DP < 10 (Preiss, 2018). These findings suggest that SSI elongates short chains, mostly DP 8-10, with SBE through their glucanotransferase reactions and create the short chains on which SSI is thought to be act [6]. It is interesting to note that the absence of SSI prevented the formation of short chains but elongated further chains with DP18. By using modified glycogen substrates, it is reported that the N-terminal mutant of maize for SSI drastically decreased the external chain length, but sharply increased SSI substrate binding ability [26]. The evidence suggested that the localization of SSI depend on starch binding by its interacting partner SSII [27]. However, it is unclear why there are short chains in wild type (WT) generated by SSI that were not extended further. A complete deficiency of SSI and SSIII in double mutant (ssI/ssIIIa) caused male sterility with opaque seeds in rice [28]. Similarly, the absence of SSI and BEIIb (branching enzyme IIb) leads to male sterility in japonica rice and this double mutant had reduced SSI level [29].
The mutant of SSII has been characterized in different crops to understand its function such as in potato tubers [18], cereal crops (endosperms of wheat [30], barley [31], rice [24], maize [32]), and in Arabidopsis leaves [33]. The observed phenotype in all crops is similar and indicates a significant change in amylopectin structure. The chain lengths of DP8 and DP18 increased and decreased in such mutants, respectively. Similarly, ssII mutants had changes in granule morphology accompanied by high amylose content and reduction in starch crystallinity [6]. Mutation for SSIIa in barley, rice and wheat have similar effects on starch structure and the amylose content but the difference in the severity of phenotypes. ssIIa mutant in rice, wheat and barley altered the structure of amylopectin which deprive the affinity of SSI to amylopectin [34]. In cereal crops, SSIIa interacts with SSI and BEII [35]. So, there can be pleiotropic effects on these enzymes due to the misfunction of SSII, making it difficult to understand the impact on phenotype due to the absence of SSII activity alone. Similarly, changes in amylopectin structure are caused by a lack of SSII activity [6]. Firstly, the recombinant rice SSII was incubated with amylopectin from the mutant ssII, which was able to promote aberrant elongation of the short chains [36]. Secondly, there was a loss of SSI activity in the ssII mutant, which caused typical ssI-type alterations in the background [37]. Thirdly, there were similar changes in amylopectin chain length distribution (CLD) in dicot plants while there is not any evidence in the formation of SSII-containing complexes [6]. The repression line for SSIIa and SSIIIa showed chalky grain appear and increased in amylose content and also decreased in viscosity in rice. In the amylopectin, there was reduction in short and long chains in grains, but number of medium chains increased. This genetically modified line nature depicted that these two genes interact each other [36].
The function of SSIII is less clear as compared to SSI and SSII. The primary role for SSIII is the formation of the B chain, elongation of cluster filling chains, and regulation of other starch synthesis enzymes. Similarly, it is also reported that SSIII also takes charge of granule initiation in the absence of SSIV. SSIII has significant activity in all plants and tissues. Analyses of ssIII mutants of maize [38] and rice [39] revealed that fewer long cluster spanning B chains (such as B2, B3, etc.) were present in mutant lines. There was also alteration in the short chains of amylopectin, indicating that SSIII also participates in the synthesis of short A and B chains [40]. These results were confirmed when compared to plants that lack SSI or SSII. It was also reported that the absence of SSII significantly affects SSIII, which results changing in the phenotypes of rice [36] and Arabidopsis [41] leaves of mutant ssII lines, suggesting partial loss of function between these two genes. The ssII/ssIII double mutant produced shorten chains with a low number of water-soluble glucans in Arabidopsis [42]. Similarly, loss of SSIIIa caused slightly reduction starch content with little rounded and smaller shape of granules in rice [43]. Additionally, the expression level of granule-bound starch synthase I (GBSSI) and ADP-glucose pyrophosphorylases increased due to absence of SSIIIa which increase amylose content [43], some of the cereal crops ss mutants are described in Table 2.
Table 2. Mutation effect on starch synthase genes in different cereal crops.
| Cereals | Amylose Content (%) | Inactivated Genes | Mutant Lines | Structural and Functional Changes in Mutant | Reference |
|---|---|---|---|---|---|
| Wheat | 22.9–32.3 | SSSII | sgp-1 | Alteration in amylopectin structure, high amylose contents | [44] |
| SSII | sgp-1, a7, a63 | Increase in short chains, decrease in starch branching enzyme | [45] | ||
| SSIIa | Increase in proportion of short chains, difference in gelatinization, retrogradation and pasting | [46] | |||
| SSIIa | svevo, semolina | Increased in dietary fiber of contents, change in total starch content, improved quality traits | [47] | ||
| SSIIa | ssIIa-Ab | Amylose contents increased 3%, cooked noodles firmness increased | [48] | ||
| SSIIa, GBSS | sw | Changes in seed size, starch granules and starch content, shrunken seed during maturity | [49] | ||
| SSIIa | abd null line | Grain properties (change in 1000 grain weight, grain size) and starch properties (fluctuation in amylose content, increased in resistant content) changed in null line | [50] | ||
| SSIV-D | e054-13, e1137 | Altered granule number/chloroplast | [51] | ||
| SSIV | e3-1-3, e1137 | Total starch and amylopectin content decreased | [52] | ||
| Rice | 15.4–25 | SSI | e7, i2-1, i2-2, i4 | Decrease in chains with DP 8 to 12, Increase in chains with DP 6 to 7 | [26] |
| SSI, SSIIIa | np | Higher amylose content, internal chain length of B2 and B3 fractions observed | [24] | ||
| SSI | ss1, isa1 | Take part in chain length distribution, outer chain elongation with little effect on branch position distribution | [53] | ||
| SSI, BEI | ss1/be1, ss1/be2b | Seed weight of mutant was higher than WT Number of short chains of amylopectin decrease, Amylose content almost same to WT |
[54] | ||
| SSI | ssI, be2b | Subtle difference in protein profile, reduced association of SSI and BEIIb in ssI mutant | [55] | ||
| SSI, SSIIa, SSIIIa | ss1L/ss2aL/ss3a | Increase amylose, decrease grain weight, increase in level of ADP-glucose pyrophosphorylases | [56] | ||
| SSII | zhonghua-15 | GC-AG intron splicing offer more variants for genetic divergence in rice | [37] | ||
| SSIIa | ss2a(em204) | SSIIa protein was totally absent in seeds, higher amylose content, Number of short chains formation increased in amylopectin | [57] | ||
| SSIIIa | ss3a-1, ss3a-2 | Chains with DP 6 to 9 and DP 16 to 19 decreased, chains with DP 10 to 15 and DP 20 to 25 increased, amylose and amylopectin content increased | [58] | ||
| SSIV-2 | allelic variation | Affected gel consistency, percent of retrogradation, | [59] | ||
| SSIIIa | flo5-1, flo5-2 | Starch granules smaller and round as compared to WT, reduced contents of long chains | [60] | ||
| SSIIIa, SSIVb | ss3a, ss4b | Produced compound type starch granules in the early stages, glucan chain length distribution identified overlapping roles for SSIIIa and SSIVb in amylopectin chain synthesis | [61] | ||
| SSIVb | Transgenic plant contains premature codons, no mRNA expression, low starch contents, dwarf phenotype | [62] | |||
| Maize | 25–30 | SSIII | dull1 | Lager clusters of chain with more branched building blocks, average cluster contained 5.4 blocks in mutant and 4.2 blocks in WT. | [63] |
| SSIII, ISA2 | du1-R4059 | Starch deficient, accumulation of phytoglycogen | [21] | ||
| SSIIa | sugary-2 | Loss of activity of endosperm specific SS, impact on the SSI and SBEIIb | [64] | ||
| SSIII | w64a | Reduced granule size, decreased the enthalpy change of starch gelatinization | [65] | ||
| Barley | 29.9–31.6 | SSII | m292, m342 | Decrease in amylopectin synthesis, pleiotropic effect on other enzymes of starch biosynthesis | [66] |
| GBSS, ISA1, SSIIa | Sex6, wax, lys5fisa1 | SSIIa mutation caused low seed weight and starch content | [67] | ||
| SSI, SSIIa, GBSS | TILLING | SSI mutant increased A and B granules, SSIIa mutant caused shrunken seed | [31] |
SSI: starch synthase I; SSII: starch synthase II; SSIII: starch synthase III; SSIV: starch synthase IV; GBSS: granule bound starch synthase; BEI: branching enzyme I; ISA1: isoamylase-type debranching enzyme 1; TILLING: Targeted induce local lesion in genomes.
2.2. Initiation of Starch Granule Formation
SSIV is involved in the initiation of the starch granule. It controls the number of starch granules in the leaves of Arabidopsis, which shows that its function is unique from other genes of the SS family [41]. The high level of starch accumulated in potato leaves is gained by a dramatic increase in the expression of the SSIV [68]. The presence of SSIV in the thylakoid membrane suggests that starch granule initiation occurs at a specific area of the chloroplast. Gene structure analysis revealed that exon and intron structure of SSIII and SSIV are highly conserved in Arabidopsis, rice, and wheat while gene structure is different from SSI, SSII, and GBSS [69].
The Arabidopsis ss4 mutant plant showed a reduction in starch granule number but had enlarged starch granules. In this case, the ADP-Glc pool is likely allocated to fewer starch granules thereby leading to considerably larger granule size in the mutant in comparison to WT (Columbia-0) plants [41]. So, this is a clear indication that the initiation of starch granules at least partially requires SSIV in Arabidopsis leaves. Interestingly, the ssI/ssII/ssIII triple mutant of Arabidopsis was able to form normal granules in the chloroplast with less starch content, highlighting the function of SSIV because granules numbers were normal in triple mutant plant [70].
References
- Polesi, L.F.; da Matta Junior, M.D.; Sarmento, S.B.S.; Canniatti-Brazaca, S.G. Starch digestibility and physicochemical and cooking properties of irradiated rice grains. Rice Sci. 2017, 24, 48–55.
- Fettke, J.; Hejazi, M.; Smirnova, J.; Höchel, E.; Stage, M.; Steup, M. Eukaryotic starch degradation: Integration of plastidial and cytosolic pathways. J. Exp. Bot. 2009, 60, 2907–2922.
- Kim, H.; Yoon, M.-R.; Chun, A.; Tai, T.H. Identification of novel mutations in the rice starch branching enzyme I gene via TILLING by sequencing. Euphytica 2018, 214, 94.
- Aoki, N.; Whitfeld, P.; Hoeren, F.; Scofield, G.; Newell, K.; Patrick, J. Three sucrose transporter genes are expressed in the developing grain of hexaploid wheat. Plant Mol. Biol. 2002, 50, 453–462.
- Kittisuban, P.; Lee, B.H.; Suphantharika, M.; Hamaker, B.R. Slow glucose release property of enzyme-synthesized highly branched maltodextrins differs among starch sources. Carbohydr. Polym. 2014, 107, 182–191.
- Pfister, B.; Zeeman, S.C. Formation of starch in plant cells. Mol. Life Sci. 2016, 73, 2781–2807.
- Cenci, U.; Nitschke, F.; Steup, M.; Minassian, B.A.; Colleoni, C.; Ball, S.G. Transition from glycogen to starch metabolism in Archaeplastida. Trends Plant Sci. 2014, 19, 18–28.
- Coutinho, P.M.; Deleury, E.; Davies, G.J.; Henrissat, B. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 2003, 328, 307–317.
- Nougué, O.; Corbi, J.; Ball, S.G.; Manicacci, D.; Tenaillon, M.I. Molecular evolution accompanying functional divergence of duplicated genes along the plant starch biosynthesis pathway. BMC Evol. Biol. 2014, 14, 103.
- Abt, M.; Pfister, B.; Sharma, M.; Eicke, S.; Bürgy, L.; Neale, I.; Seung, D.; Zeeman, S.C. Starch Synthase 5, a Noncanonical Starch Synthase-Like Protein, Promotes Starch Granule Initiation in Arabidopsis. Plant Cell. 2020.
- Zhu, F. Interactions between starch and phenolic compound. Trends Food Sci. Technol. 2015, 43, 129–143.
- Gous, P.W.; Fox, G.P. Amylopectin synthesis and hydrolysis—Understanding isoamylase and limit dextrinase and their impact on starch structure on barley (Hordeum vulgare) quality. J. Trends Food Sci. 2017, 62, 23–32.
- Pandey, M.K.; Rani, N.S.; Madhav, M.S.; Sundaram, R.; Varaprasad, G.; Sivaranjani, A.; Bohra, A.; Kumar, G.R.; Kumar, A. Different isoforms of starch-synthesizing enzymes controlling amylose and amylopectin content in rice (Oryza sativa L.). Biotechnol. Adv. 2012, 30, 1697–1706.
- Pan, X.; Yan, H.; Li, M.; Wu, G.; Jiang, H. Evolution of the genes encoding starch synthase in Sorghum and common wheat. Mol. Plant Breed. 2011, 2, 60–67.
- Wang, Z.; Li, W.; Qi, J.; Shi, P.; Yin, Y. Starch accumulation, activities of key enzyme and gene expression in starch synthesis of wheat endosperm with different starch contents. J. Food Sci. Technol. 2014, 51, 419–429.
- Qu, J.; Ma, C.; Feng, J.; Xu, S.; Wang, L.; Li, F.; Li, Y.; Zhang, R.; Zhang, X.; Xue, J. Transcriptome dynamics during maize endosperm development. PLoS ONE 2016, 3, e0163814.
- Crofts, N.; Abe, N.; Oitome, N.F.; Matsushima, R.; Hayashi, M.; Tetlow, I.J. Amylopectin biosynthetic enzymes from developing rice seed form enzymatically active protein complexes. J. Exp. Bot. 2015, 66, 4469–4482.
- Edwards, A.; Fulton, D.C.; Hylton, C.M.; Jobling, S.A.; Gidley, M.; Rössner, U. A combined reduction in activity of starch synthases II and III of potato has novel effects on the starch of tubers. Plant J. 1999, 17, 251–261.
- Cuesta-Seijo, J.A.; Nielsen, M.M.; Ruzanski, C.; Krucewicz, K.; Beeren, S.R.; Rydhal, M.G. In vitro biochemical characterization of all barley endosperm starch synthases. Front. Plant Sci. 2016, 6, 1265.
- Huang, B.; Keeling, P.L.; Hennen-Bierwagen, T.A.; Myers, A.M. Comparative in vitro analyses of recombinant maize starch synthases SSI, SSIIa, and SSIII reveal direct regulatory interactions and thermosensitivity. Arch. Biochem. Biophys. 2016, 596, 63–72.
- Lin, Q.; Qian, S.; Li, C.; Pan, H.; Wu, Z.; Liu, G. Synthesis, flocculation and adsorption performance of amphoteric starch. Carbohydr. Polym. 2012, 90, 275–283.
- Tetlow, I.J.; Emes, M.J. Starch biosynthesis in the developing endosperms of grasses and cereals. Agronomy 2017, 7, 81.
- Šárka, E.; Dvořáček, V. New processing and applications of waxy starch (a review). J. Food Eng. 2017, 206, 77–87.
- Hanashiro, I.; Higuchi, T.; Aihara, S.; Nakamura, Y.; Fujita, N. Structures of starches from rice mutants deficient in the starch synthase isozyme SSI or SSIIIa. J. Biomacromol. 2011, 12, 1621–1628.
- Brust, H.; Lehmann, T.; D’Hulst, C.; Fettke, J. Analysis of the functional interaction of Arabidopsis starch synthase and branching enzyme isoforms reveals that the cooperative action of SSI and BEs results in glucans with polymodal chain length distribution similar to amylopectin. PLoS ONE 2014, 9, e102364.
- Fujita, N.; Yoshida, M.; Asakura, N.; Ohdan, T.; Miyao, A.; Hirochika, H.; Nakamura, Y. Function and characterization of starch synthase I using mutants in rice. Plant Physiol. 2006, 140, 1070–1084.
- Wilkens, C.; Svensson, B.; Møller, M.S. Functional roles of starch binding domains and surface binding sites in enzymes involved in starch biosynthesis. Front. Plant Sci. 2018, 9, 1652.
- Fujita, N.; Satoh, R.; Hayashi, A.; Kodama, M.; Itoh, R.; Aihara, S.; Nakamura, Y. Starch biosynthesis in rice endosperm requires the presence of either starch synthase I or IIIa. J. Exp. Bot. 2011, 62, 4819–4831.
- Nakata, M.; Miyashita, T.; Kimura, R.; Nakata, Y.; Takagi, H.; Kuroda, M.; Yamaguchi, T.; Umemoto, T.; Yamakawa, H. MutMapPlus identified novel mutant alleles of a rice starch branching enzyme II b gene for fine-tuning of cooked rice texture. Plant Biotechnol. J. 2018, 16, 111–123.
- Guzmán, C.; Alvarez, J.B. Wheat waxy proteins: Polymorphism, molecular characterization and effects on starch properties. Theor. Appl. Genet. 2016, 129, 1–16.
- Sparla, F.; Falini, G.; Botticella, E.; Pirone, C.; Talamè, V.; Bovina, R.; Salvi, S.; Tuberosa, R.; Sestili, F.; Trost, P. New starch phenotypes produced by TILLING in barley. PLoS ONE 2014, 9, e107779.
- Niu, L.; Ding, H.; Zhang, J.; Wang, W. Proteomic analysis of starch biosynthesis in maize seeds. Starch Starke 2019, 71, 1800294.
- Szydlowski, N.; Ragel, P.; Hennen-Bierwagen, T.A.; Planchot, V.; Myers, A.M.; Mérida, A.; d’Hulst, C.; Wattebled, F. Integrated functions among multiple starch synthases determine both amylopectin chain length and branch linkage location in Arabidopsis leaf starch. J. Exp. Bot. 2011, 62, 4547–4559.
- Luo, J.; Ahmed, R.; Kosar-Hashemi, B.; Larroque, O.; Butardo, V.M.; Tanner, G.J.; Colgrave, M.L.; Upadhyaya, N.M.; Tetlow, I.J.; Emes, M.J.; et al. The different effects of starch synthase IIa mutations or variation on endosperm amylose content of barley, wheat and rice are determined by the distribution of starch synthase I and starch branching enzyme IIb between the starch granule and amyloplast stroma. Theor. Appl. Genet. 2015, 128, 1407–1419.
- Subasinghe, R.M.; Liu, F.; Polack, U.C.; Lee, E.A.; Emes, M.J.; Tetlow, I.J. Multimeric states of starch phosphorylase determine protein–protein interactions with starch biosynthetic enzymes in amyloplasts. Plant Physiol. Biochem. 2014, 83, 168–179.
- Zhang, G.; Cheng, Z.; Zhang, X.; Guo, X.; Su, N.; Jiang, L.; Mao, L.; Wan, J. Double repression of soluble starch synthase genes SSIIa and SSIIIa in rice (Oryza sativa L.) uncovers interactive effects on the physicochemical properties of starch. Genome 2011, 54, 448–459.
- Chen, C.; Gao, S.; Sun, Q.; Tang, Y.; Han, Y.; Zhang, J.; Li, Z. Induced splice site mutation generates alternative intron splicing in starch synthase II (SSII) gene in rice. Biotechnol. Equip. 2017, 31, 1093–1099.
- Zhu, F.; Bertoft, E.; Seetharaman, K. Distribution of branches in whole starches from maize mutants deficient in starch synthase III. J. Agric. Food Chem. 2014, 62, 4577–4583.
- Takahashi, T.; Fujita, N. Thermal and rheological characteristics of mutant rice starches with widespread variation of amylose content and amylopectin structure. Food Hydrocoll. 2017, 62, 83–93.
- Lin, Q.; Huang, B.; Zhang, M.; Zhang, X.; Rivenbark, J.; Lappe, R.L.; James, M.G.; Myers, A.M.; Hennen-Bierwagen, T.A. Functional interactions between starch synthase III and isoamylase-type starch-debranching enzyme in maize endosperm. Plant Physiol. 2012, 158, 679–692.
- Santelia, D.; Zeeman, S.C. Progress in Arabidopsis starch research and potential biotechnological applications. Curr. Opin. Biotechnol. 2011, 22, 271–280.
- Zhang, X.; Szydlowski, N.; Delvallé, D.; d’Hulst, C.; James, M.G.; Myers, A.M. Overlapping functions of the starch synthases SSII and SSIII in amylopectin biosynthesis in Arabidopsis. BMC Plant Biol. 2008, 8, 96.
- Hayashi, M.; Kodama, M.; Nakamura, Y.; Fujita, N. Thermal and pasting properties, morphology of starch granules, and crystallinity of endosperm starch in the rice SSI and SSIIIa double-mutant. J. Appl. Glycosci. 2015, 62, 81–86.
- Yamamori, M.; Fujita, S.; Hayakawa, K.; Matsuki, J.; Yasui, T. Genetic elimination of a starch granule protein, SGP-1, of wheat generates an altered starch with apparent high amylose. Theor. Appl. Genet. 2000, 101, 21–29.
- Kosar-Hashemi, B.; Li, Z.; Larroque, O.; Regina, A.; Yamamori, M.; Morell, M.K.; Rahman, S. Multiple effects of the starch synthase II mutation in developing wheat endosperm. Funct. Plant Biol. 2007, 34, 431–438.
- Vrinten, P.L.; Shimbata, T.; Yanase, M.; Sunohara, A.; Saito, M.; Inokuma, T.; Takiya, T.; Takaha, T.; Nakamura, T. Properties of a novel type of starch found in the double mutant “sweet wheat”. Carbohydr. Polym. 2012, 89, 1250–1260.
- Botticella, E.; Sestili, F.; Ferrazzano, G.; Mantovani, P.; Cammerata, A.; D’Egidio, M.G.; Lafiandra, D. The impact of the SSIIa null mutations on grain traits and composition in durum wheat. Breed. Sci. 2016, 16, 4.
- Martin, J.M.; Hogg, A.C.; Hofer, P.; Manthey, F.A.; Giroux, M.J. Impacts of SSIIa-A null allele on durum wheat noodle quality. Cereal Chem. 2014, 91, 176–182.
- Shimbata, T.; Ai, Y.; Fujita, M.; Inokuma, T.; Vrinten, P.; Sunohara, A.; Saito, M.; Takiya, T.; Jane, J.L.; Nakamura, T. Effects of homoeologous wheat starch synthase IIa genes on starch properties. J. Agric. Food Chem. 2012, 60, 12004–12010.
- Konik-Rose, C.; Thistleton, J.; Chanvrier, H.; Tan, I.; Halley, P.; Gidley, M.; Kosar-Hashemi, B.; Wang, H.; Larroque, O.; Ikea, J.; et al. Effects of starch synthase IIa gene dosage on grain, protein and starch in endosperm of wheat. Theor. Appl. Genet. 2007, 115, 1053.
- Guo, H.; Liu, Y.; Li, X.; Yan, Z.; Xie, Y.; Xiong, H.; Zhao, L.; Gu, J.; Zhao, S.; Liu, L. Novel mutant alleles of the starch synthesis gene TaSSIVb-D result in the reduction of starch granule number per chloroplast in wheat. BMC Genom. 2017, 18, 358.
- Zhang, S.; Guo, H.; Irshad, A.; Xie, Y.; Zhao, L.; Xiong, H.; Gu, J.; Zhao, S.; Ding, Y.; Liu, L. The synergistic effects of TaAGP. L-B1 and TaSSIVb-D mutations in wheat lead to alterations of gene expression patterns and starch content in grain development. PLoS ONE 2019, 14, e0223783.
- Fujita, N.; Hanashiro, I.; Toyosawa, Y.; Nakamura, Y. Functional study of rice starch synthase I (SSI) by using double mutant with lowered activities of SSI and isoamylase1. J. Appl. Glycosci. 2013, 60, 45–51.
- Abe, N.; Asai, H.; Yago, H.; Oitome, N.F.; Itoh, R.; Crofts, N.; Nakamura, Y.; Fujita, N. Relationships between starch synthase I and branching enzyme isozymes determined using double mutant rice lines. BMC Plant Biol. 2014, 14, 80.
- Crofts, N.; Iizuka, Y.; Abe, N.; Miura, S.; Kikuchi, K.; Matsushima, R.; Fujita, N. Rice mutants lacking starch synthase I or branching enzyme IIb activity altered starch biosynthetic protein complexes. Front. Plant Sci. 2018, 9, 1817.
- Hayashi, M.; Crofts, N.; Oitome, N.F.; Fujita, N. Analyses of starch biosynthetic protein complexes and starch properties from developing mutant rice seeds with minimal starch synthase activities. BMC Plant Biol. 2018, 18, 59.
- Miura, S.; Crofts, N.; Saito, Y.; Hosaka, Y.; Oitome, N.F.; Watanabe, T.; Kumamaru, T.; Fujita, N. Starch synthase IIa-deficient mutant rice line produces endosperm starch with lower gelatinization temperature than japonica rice cultivars. Front. Plant Sci. 2018, 9, 645.
- Fujita, N.; Yoshida, M.; Kondo, T.; Saito, K.; Utsumi, Y.; Tokunaga, T.; Nishi, A.; Satoh, H.; Park, J.-H. Characterization of SSIIIa-deficient mutants of rice: The function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol. 2007, 144, 2009–2023.
- Xu, L.; You, H.; Zhang, O.; Xiang, X. Genetic Effects of Soluble Starch Synthase IV-2 and It with ADPglucose Pyrophorylase Large Unit and Pullulanase on Rice Qualities. Rice 2020, 13, 46.
- Ryoo, N.; Yu, C.; Park, C.-S.; Baik, M.-Y.; Park, I.M.; Cho, M.-H.; Bhoo, S.H.; An, G.; Hahn, T.-R.; Jeon, J.-S. Knockout of a starch synthase gene OsSSIIIa/Flo5 causes white-core floury endosperm in rice (Oryza sativa L.). Plant Cell Rep. 2007, 26, 1083–1095.
- Toyosawa, Y.; Kawagoe, Y.; Matsushima, R.; Crofts, N.; Ogawa, M.; Fukuda, M.; Kumamaru, T.; Okazaki, Y.; Kusano, M.; Saito, K. Deficiency of starch synthase IIIa and IVb alters starch granule morphology from polyhedral to spherical in rice endosperm. Plant Physiol. 2016, 170, 1255–1270.
- Jung, Y.-J.; Nogoy, F.M.; Lee, S.-K.; Cho, Y.-G.; Kang, K.-K. Application of ZFN for site directed mutagenesis of rice SSIVa gene. Biotechnol. Bioproc. Eng. 2018, 23, 108–115.
- Zhu, F.; Bertoft, E.; Källman, A.; Myers, A.M.; Seetharaman, K. Molecular structure of starches from maize mutants deficient in starch synthase III. J. Agric. Food Chem. 2013, 61, 9899–9907.
- Liu, F.; Ahmed, Z.; Lee, E.A.; Donner, E.; Liu, Q.; Ahmed, R.; Morell, M.K.; Emes, M.J.; Tetlow, I.J. Allelic variants of the amylose extender mutation of maize demonstrate phenotypic variation in starch structure resulting from modified protein–protein interactions. J. Exp.Bot. 2012, 63, 1167–1183.
- Zhu, F.; Bertoft, E.; Li, G. Morphological, thermal, and rheological properties of starches from maize mutants deficient in starch synthase III. J. Agric. Food Chem. 2016, 64, 6539–6545.
- Morell, M.K.; Kosar-Hashemi, B.; Cmiel, M.; Samuel, M.S.; Chandler, P.; Rahman, S.; Buleon, A.; Batey, I.L.; Li, Z. Barley sex6 mutants lack starch synthase IIa activity and contain a starch with novel properties. Plant J. 2003, 34, 173–185.
- Howard, T.P.; Fahy, B.; Leigh, F.; Howell, P.; Powell, W.; Greenland, A.; Trafford, K.; Smith, A.M. Use of advanced recombinant lines to study the impact and potential of mutations affecting starch synthesis in barley. J. Cereal Sci. 2014, 59, 196–202.
- Ezquer, I.; Li, J.; Ovecka, M.; Baroja-Fernández, E.; Muñoz, F.J.; Montero, M.; Díaz de Cerio, J.; Hidalgo, M.; Sesma, M.T.; Bahaji, A. Microbial volatile emissions promote accumulation of exceptionally high levels of starch in leaves in mono-and dicotyledonous plants. Plant Cell Physiol. 2010, 51, 1674–1693.
- Irshad, A.; Guo, H.; Zhang, S.; Gu, J.; Zhao, L.; Xie, Y.; Xiong, H.; Zhao, S.; Ding, Y.; Ma, Y. EcoTILLING reveals natural allelic variations in starch synthesis key gene TaSSIV and Its haplotypes associated with higher thousand grain weight. Genes 2019, 10, 307.
- Gámez-Arjona, F.M.; Raynaud, S.; Ragel, P.; Mérida, Á. Starch synthase 4 is located in the thylakoid membrane and interacts with plastoglobule-associated proteins in Arabidopsis. Plant J. 2014, 80, 305–316.
More
Information
Subjects:
Agriculture, Dairy & Animal Science
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.2K
Revisions:
2 times
(View History)
Update Date:
27 Oct 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




