
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Bo Zhou | + 1995 word(s) | 1995 | 2021-10-19 07:58:36 | | | |
| 2 | Catherine Yang | Meta information modification | 1995 | 2021-10-25 04:12:48 | | |
Video Upload Options
Anthocyanins are natural pigments with antioxidant effects that exist in various fruits and vegetables. The accumulation of anthocyanins is induced by environmental signals and regulated by transcription factors in plants. Numerous evidence has indicated that among the environmental factors, light is one of the most signal regulatory factors involved in the anthocyanin biosynthesis pathway.
1. Introduction
Plant growth and development are greatly affected by the environment. Anthocyanin production is affected by various environmental factors such as light, low temperature, drought, and salinity [1][2][3]. Light is a particularly important environmental factor inducing anthocyanin accumulation in plants. Different types of anthocyanin are widely distributed in various plant tissues such as flowers, fruits, stems, leaves, and underground tubers [4]. The anthocyanin accumulation in these plant tissues is shown as light-dependent or light-independent biosynthesis. The different light quality and intensity are perceived by receptors or signal factors to transduce them into downstream transcription factors [5]. Next, metabolites are synthesized by enzymes encoded by structural genes that are regulated by transcription factors to respond to light. Anthocyanins belonging to the flavonoids are important secondary metabolites to adapt to harmful environmental stress in plants.
Anthocyanins are universal water-soluble flavonoid pigments that are responsible for the widest color of leaves, petals, fruits, seeds, stems, and tubers of plants (Liu et al. , 2021) [6]. Moreover, they have been shown to play a beneficial role in the visual activity, cancer, heart disease, and age-related neurodegenerative disorders [7], although no effects or even a negative effect on health-related parameters have also been reported [6][8]. Anthocyanins consist of an anthocyanidin backbone with sugar and acyl conjugates while anthocyanidins are composed of two aromatic benzene rings and an oxygenated heterocycle [9][10]. Due to the number of hydroxyl groups and glycosyl groups in the rings, anthocyanins have potent antioxidant activity to scavenge free radicals and reactive oxygen species (ROS) during environmental stresses [6]. Therefore, anthocyanins have protective effects during plant development through absorbing excess UV light, preventing lipid peroxidation and suppressing the activity of ROS. Then, plants have evolved to the biosynthetic pathways of anthocyanins to resist various abiotic stresses including UV irradiation, drought, high salinity, and low temperature [11].
2. Anthocyanins Accumulation in Plants
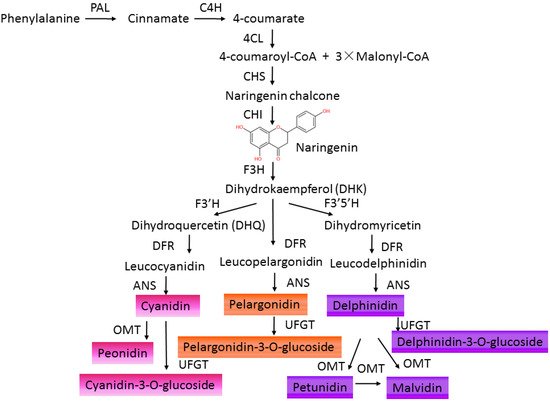
In the natural environment, anthocyanins participate in the formation of multicolors in fruits, flowers, leaves, even roots of plants [7][12]. Different kinds of anthocyanins modified by methylation and hydroxylation at different positions of C6-C3-C6 carbon skeleton structures decide different colors varying from orange, red, and purple to blue in plants [7][13] ( Table 1 ). The physiological role of anthocyanin in plants consists of defending against viral, bacterial, and fungicidal activities, absorbing excess visible and UV light irradiance, attracting pollinators and seed dispersers, and scavenging excess ROS under abiotic stresses [6][11][14]. Anthocyanins are synthesized in the endoplasmic reticulum and transported to accumulate in the vacuoles of a wide range of cells and tissues in both the vegetative and reproductive organs of plants [14]. The synthesis of anthocyanins in plants is controlled by structural genes and can be divided into four stages [15][16]. The early stage is from phenylalanine to 4-coumaryl CoA sequentially catalyzed by phenylalanine ammonialyase (PAL), cinnamate 4-hydroxylase (C4H), and 4-coumarate-CoA ligase (4CL). Then, 4-coumaroyl CoA is converted to dihydrokaempferol (DHK), which is regulated by chalcone synthase (CHS), chalcone isomerase (CHI), and flavanone 3-hydroxylase (F3H). DHK can also be converted to dihydroquercetin (DHQ) by flavonoid 3′-hydroxylase (F3′H) or catalyzed by flavonoid 3′5′-hydroxylase (F3′5′H) to form dihydromyricetin. Next, DHK, DHQ, and dihydromyricetin are separately converted to leucopelargonidin, leucocyanidin, and leucodelphinidin by dihydroflavonol 4-reductase (DFR). Finally, these leucoanthocyanidins are catalyzed by anthocyanidin synthase/leucoanthocyanidin dioxygenase (ANS/LDOX) to form colored anthocyanidins (pelargonidin, cyanidin, delphinidin) [17]. Then, these anthocyanidins can be further modified by UDP-glucose flavonoid glucosyltransferase (UFGT), acetylase, O-methyltransferase (OMT), and anthocyanin transferase (AT) [18] to produce stable and water-soluble pigments ( Figure 1 ). These flavonoid products vary in different plants due to the evolution of anthocyanin metabolism under environmental selective pressures [19]. Anthocyanins show potent antioxidant activities and are effective scavengers of ROS in vitro [20][21]. In addition, anthocyanins are involved in regulating ROS-induced signaling responding to environmental cues [22][23][24]. Anthocyanins may be regulators of the ROS-signaling network due to their ability to interact with protein and to enhance the activity of protein through the ROS reaction [14][25][26]. However, the functional roles of flavonoids in plants remain indistinct not only for their numerous varieties, but also for their different influences in different plant species. According to different properties of solubility, light absorption, and distribution patterns in various parts of the plant, anthocyanins have been cataloged numerically [27], and the composition profile of anthocyanins varied under different stresses [28][29]. The biological functions of different types of anthocyanins still need to be identified in various plants. Moreover, the roles of family genes, encoded enzymes of the anthocyanin biosynthetic pathway, also need to be clarified [30][31][32].
| Chemical Structure | Substitution | Anthocyanidin Name | Color | λmax in the Visible Spectrum | |
|---|---|---|---|---|---|
| R1 | R2 | ||||
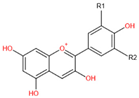 |
H | H | Pelargonidin (Pg) | Orange | 494 nm |
| OH | H | Cyanidin (Cy) | Orange-red | 504 nm | |
| OH | OH | Delphinidin (Dp) | Blue-red | 508 nm | |
| OCH3 | H | Peonidin (Pn) | Orange-red | 506 nm | |
| OCH3 | OH | Petunidin (Pt) | Blue-Red | 508 nm | |
| OCH3 | OCH3 | Malvidin (Mv) | Blue-red | 510 nm | |
3. Regulatory Pathway of Anthocyanin Biosynthesis
According to the research of Arabidopsis , it is well known that early anthocyanin biosynthetic genes ( CHS , CHI , F3H , F3′H , and FLS ) are regulated by R2R3-MYB transcription factors MYB11, MYB12, MYB111, and MYB75 while the late biosynthetic genes ( DFR , ANS / LDOX , UFGT ) are regulated by the MBW complex (R2R3-MYBs such as MYB75, MYB90, MYB113, and MYB114, bHLH, and WD40) [33][34][35]. In addition to MYB-bHLH-WDR transcription factors, other regulatory proteins have also been identified to be involved in anthocyanin biosynthesis in plants including members of the NAC transcription family [36][37], MADS-box proteins [38][39][40], bZIP transcription factors [41][42][43], WRKY transcription factors [44], and SPL transcription factors [45][46]. These transcription factors take positive or negative roles in regulating the expression of structural anthocyanin biosynthesis genes and determine the anthocyanin accumulation in developmental and environmental regulation.
Among these transcription factors, MYB transcription factors play a significant role in regulating anthocyanin biosynthesis. Many anthocyanin-related R2R3-MYB factors have been identified from many plants such as Arabidopsis [33], grape [47], Gerbera [48], eggplant [49], tomato [49], populous [50], apple [51][52], etc. Most of these are R2R3-MYB proteins with the motif [D/E]LX2[R/K]X3LX6LX3R in the R3 domain, which is necessary to interact with bHLH acting as activators to increase anthocyanin accumulation [53]. In Arabidopsis , MYB75 (PAP1), MYB90 (PAP2), MYB113 (PAP3), and MYB114 (PAP4) show very high conserved sequences, and their overexpression increases the accumulation of anthocyanin through TTG1- and bHLH-dependent regulating late anthocyanin pathway genes [33]. In addition, the transcription level of PAP1 is higher than that of the other three MYB genes in young seedlings of Arabidopsis . In AtMYB75 transgenic tomato plants, anthocyanin production was also induced in tomato plants [54]. Moreover, overexpressing a single SlMYB75 TF can lead to abundant anthocyanin accumulation in both vegetative and reproductive organs of tomatoes [55] and another MYB TF SlAN2-like was also identified to be responsible for the Aft phenotype (anthocyanin fruit). In blueberry, VcMYBL1 has been identified to interact with VcbHLHL1 to promote anthocyanin biosynthesis [56]. In apple, MdMYB3 is also characterized to activate transcriptional flavonoid pathway genes and regulate the accumulation of anthocyanin in the skin of apple fruits [57]. SmMYB1 isolated in eggplant can interact with a heterologous bHLH, but the C-terminal domain in SmMYB1 is essential for transcriptional activation of anthocyanin genes [58].
These R2R3 MYBs have conserved R2R3 domain sequences, which determine anthocyanin pathway specificity. For example, in Arabidopsis , PAP4 activates the transcription of UFGT , which can produce anthocyanins from anthocyanidins, while TT2 regulates the expression of ANR , which is specific to proanthocyanin biosynthesis [59]. Alteration of individual amino acids of the R2R3 domain in PAP4 and TT2 can lead to changes in activation of the ANR and UFGT promoter. The amino-acid change in TT2 (Gly 39 →Arg) can switch TT2 specificity toward the anthocyanin pathway while the amino-acid mutant (Arg 39 → Gly) and exchange of motif (Ala-Asn-Asp-Val → Asp-Asn-Glu-Ile 90−93 ) in PAP4 switches PAP4 specificity toward the PA pathway [59]. The orthologs VvMYBA2 and VvMYBPA2 in grapevine also have similar amino acids for pathway specificity [59]. Therefore, the anthocyanin pathway specificity is determined by the amino acid composition of the R2R3 domain in MYB transcription factors and the cis-elements in the promoters of anthocyanin biosynthetic genes. Due to the conservation of R2R3-MYB, regulatory differences are determined by promoter divergence rather than the divergence of the regulators [59][60]. Then, it is not difficult to understand that MYB transcription factors particularly regulate the expression of different anthocyanin pathway genes in various plants. In Arabidopsis , MYB11, MYB12, and MYB111 activate promoters of early genes ( CHS , CHI , F3H , and FLS ) but not of late genes ( F3′H and DFR ) [61][62]. However, the expression of PAL1 and CHS remains unchanged in MYB75, MYB90, MYB113, and MYB114 RNAi seedlings, and the transcription for the late genes DFR , F3′H , UGT75C1 , LDOX , and GST12 is reduced [33]. In apple fruit, MdMYB114 also promotes anthocyanin accumulation by directly controlling the expression of late genes MdANS , MdUFGT , and MdGST [52], but in turnip ( Brassica rapa ), BrMYB75 can bind the promoter of early gene CHS to regulate anthocyanin biosynthesis [30]. Moreover, overexpression of MdMYB90-like in apple bud can induce the expression of both early and late genes including MdCHS , MdCHI , MdANS , and MdUFGT [51]. In tomatoes, two anthocyanin-related R2R3-MYB factors take different tissue-specific regulatory roles in anthocyanin accumulation. One is SlAN2/SlMYB75, which is as a positive regulator of anthocyanin biosynthesis in vegetative tissues of tomato plants including hypocotyls, cotyledons, stems, and leaves [63], and the other is SlAN2-like /Aft, which determines anthocyanins in the fruits of tomato plants [64][65].
As cofactors of MYB activators and repressors, bHLH proteins incorporating the MYB-interacting region, WD40/AD domain, basic helix–loop–helix domain, and ACT domain are responsible for MYB interaction and promoter binding of anthocyanin biosynthetic genes [66]. The bHLH family has been divided into 26 subfamilies [67] and the IIIf subfamily bHLH members are involved in both flavonoid biosynthesis and trichome formation [68]. The binding of the bHLH proteins with WD40 protein can be modulated by different R2R3 MYB proteins. In Arabidopsis , three bHLH TFs, GL3, EGL3, and TT8, have been shown to interact with R2R3-MYB proteins and regulate flavonoid biosynthesis [33][69]. AtTT8 can interact with AtTTG1 to form stable MBW complexes with different R2R3 MYBs while AtGL3 and AtEGL3 show competitive complex formation with some R2R3 MYBs [70]. The AtTTG1–bHLH interactions are modulated positively and negatively through the addition of R2R3 MYB proteins in Arabidopsis , [70]. PhAN1 and PhJAF13 in petunia have also been identified to be involved in anthocyanin [71][72]. In Solanaceous plants, AN1 directly regulates the expression of biosynthetic genes, whereas JAF13 can regulate the transcription of AN1 [73]. Then, the IIIf subfamily bHLHs can also be further divided into JAF13 clade and AN1 clade [72]. In the petunia hybrid, the interaction of PhAN1 with PhAN11 is promoted by the addition of the phAN4 R2R3 MYB protein, while the interaction of PhJAF13 with PhAN11 is significantly reduced in the presence of R2R3 MYB proteins [70]. Therefore, in different species, even in different tissues of the same plant, the functions of the bHLHs and MBW complexes vary. For example, AcMYB123 and AcbHLH42 can activate the function of the promoters of AcF3GT1 and AcANS and be involved in the spatiotemporal regulation of anthocyanin biosynthesis, specifically in the inner pericarp of kiwifruit [74]. MdbHLH3 can also bind to the promoters of MdDFR , MdUFGT , and MdMYB1 to activate their expression, which is involved in anthocyanin biosynthesis in apples. The individual combinations of bHLH and R2R3 MYB proteins regulate the expression of different anthocyanin synthetic genes and determine the spatiotemporal pigment accumulation in plants.
References
- Kumar, A.A.; Mishra, P.; Kumari, K.; Panigrahi, K.C. Environmental stress influencing plant development and flowering. Front. Biosci. 2012, 4, 1315–1324.
- Chen, Z.H.; Soltis, D.E. Evolution of environmental stress responses in plants. Plant Cell Environ. 2020, 43, 2827–2831.
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483.
- Saigo, T.; Wang, T.; Watanabe, M.; Tohge, T. Diversity of anthocyanin and proanthocyanin biosynthesis in land plants. Curr. Opin. Plant Biol. 2020, 55, 93–99.
- Kong, S.G.; Okajima, K. Diverse photoreceptors and light responses in plants. J. Plant Res. 2016, 129, 111–114.
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin Biosynthesis and Degradation Mechanisms in Solanaceous Vegetables: A Review. Front. Chem. 2018, 6, 52.
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779.
- Smeriglio, A.; Barreca, D.; Bellocco, E.; Trombetta, D. Chemistry, Pharmacology and Health Benefits of Anthocyanins. Phytother. Res. 2016, 30, 1265–1286.
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749.
- Stommel, J.R.; Lightbourn, G.J.; Winkel, B.S.; Griesbach, R.J. Transcription Factor Families Regulate the Anthocyanin Biosynthetic Pathway in Capsicum annuum. J. Am. Soc. Hortic. Sci. 2009, 134, 244–251.
- Naing, A.H.; Kim, C.K. Abiotic stress-induced anthocyanins in plants: Their role in tolerance to abiotic stresses. Physiol. Plantarum 2021, 172, 1711–1723.
- Mattioli, R.; Francioso, A.; Mosca, L.; Silva, P. Anthocyanins: A Comprehensive Review of Their Chemical Properties and Health Effects on Cardiovascular and Neurodegenerative Diseases. Molecules 2020, 25, 3809.
- Liu, H.; Liu, Z.; Wu, Y.; Zheng, L.; Zhang, G. Regulatory Mechanisms of Anthocyanin Biosynthesis in Apple and Pear. Int. J. Mol. Sci. 2021, 22, 8441.
- Landi, M.; Tattini, M.; Gould, K.S. Multiple functional roles of anthocyanins in plant-environment interactions. Environ. Exp. Bot. 2015, 119, 4–17.
- Kitamura, S.; Shikazono, N.; Tanaka, A. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J. 2004, 37, 104–114.
- Routaboul, J.M.; Dubos, C.; Beck, G.; Marquis, C.; Bidzinski, P.; Loudet, O.; Lepiniec, L. Metabolite profiling and quantitative genetics of natural variation for flavonoids in Arabidopsis. J. Exp. Bot. 2012, 63, 3749–3764.
- Chaves-Silva, S.; dos Santos, A.L.; Chalfun, A.; Zhao, J.; Peres, L.E.P.; Benedito, V.A. Understanding the genetic regulation of anthocyanin biosynthesis in plants—Tools for breeding purple varieties of fruits and vegetables. Phytochemistry 2018, 153, 11–27.
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493.
- Serna, L.; Martin, C. Trichomes: Different regulatory networks lead to convergent structures. Trends Plant Sci. 2006, 11, 274–280.
- Ferreres, F.; Figueiredo, R.; Bettencourt, S.; Carqueijeiro, I.; Oliveira, J.; Gil-Izquierdo, A.; Pereira, D.M.; Valentao, P.; Andrade, P.B.; Duarte, P.; et al. Identification of phenolic compounds in isolated vacuoles of the medicinal plant Catharanthus roseus and their interaction with vacuolar class III peroxidase: An H2O2 affair? J. Exp. Bot. 2011, 62, 2841–2854.
- Juadjur, A.; Mohn, C.; Schantz, M.; Baum, M.; Winterhalter, P.; Richling, E. Fractionation of an anthocyanin-rich bilberry extract and in vitro antioxidative activity testing. Food Chem. 2015, 167, 418–424.
- Heine, G.F.; Hernandez, J.M.; Grotewold, E. Two cysteines in plant R2R3 MYB domains participate in REDOX-dependent DNA binding. J. Biol. Chem. 2004, 279, 37878–37885.
- Taylor, L.P.; Grotewold, E. Flavonoids as developmental regulators. Curr. Opin. Plant Biol. 2005, 8, 317–323.
- Brunetti, C.; Di Ferdinando, M.; Fini, A.; Pollastri, S.; Tattini, M. Flavonoids as Antioxidants and Developmental Regulators: Relative Significance in Plants and Humans. Int. J. Mol. Sci. 2013, 14, 3540–3555.
- Payne, J.L.; Moore, J.H.; Wagner, A. Robustness, Evolvability, and the Logic of Genetic Regulation. Artif. Life 2014, 20, 111–126.
- Lin, Y.; Fan, L.; He, J.; Wang, Z.; Yin, Y.; Cheng, Y.; Li, Z. Anthocyanins contribute to fruit defense against postharvest green mold. Postharvest Biol. Technol. 2021, 181, 111661.
- Luo, J.; Nishiyama, Y.; Fuell, C.; Taguchi, G.; Elliott, K.; Hill, L.; Tanaka, Y.; Kitayama, M.; Yamazaki, M.; Bailey, P.; et al. Convergent evolution in the BAHD family of acyl transferases: Identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana. Plant J. 2007, 50, 678–695.
- Kovinich, N.; Kayanja, G.; Chanoca, A.; Riedl, K.; Otegui, M.S.; Grotewold, E. Not all anthocyanins are born equal: Distinct patterns induced by stress in Arabidopsis. Planta 2014, 240, 931–940.
- Kovinich, N.; Kayanja, G.; Chanoca, A.; Otegui, M.S.; Grotewold, E. Abiotic stresses induce different localizations of anthocyanins in Arabidopsis. Plant Signal. Behav. 2015, 10, e1027850.
- Zhou, B.; Wang, Y.; Zhan, Y.G.; Li, Y.H.; Kawabata, S. Chalcone Synthase Family Genes Have Redundant Roles in Anthocyanin Biosynthesis and in Response to Blue/Uv-a Light in Turnip (Brassica Rapa; Brassicaceae). Am. J. Bot. 2013, 100, 2458–2467.
- Li, Z.; Vickrey, T.L.; McNally, M.G.; Sato, S.J.; Clemente, T.E.; Mower, J.P. Assessing Anthocyanin Biosynthesis in Solanaceae as a Model Pathway for Secondary Metabolism. Genes 2019, 10, 559.
- Wu, X.X.; Zhang, S.M.; Liu, X.H.; Shang, J.; Zhang, A.D.; Zhu, Z.W.; Zha, D.S. Chalcone synthase (CHS) family members analysis from eggplant (Solanum melongena L.) in the flavonoid biosynthetic pathway and expression patterns in response to heat stress. PLoS ONE 2020, 15, e0226537.
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827.
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581.
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185.
- Wang, J.F.; Lian, W.R.; Cao, Y.Y.; Wang, X.Y.; Wang, G.L.; Qi, C.D.; Liu, L.; Qin, S.J.; Yuan, X.W.; Li, X.S.; et al. Overexpression of BoNAC019, a NAC transcription factor from Brassica oleracea, negatively regulates the dehydration response and anthocyanin biosynthesis in Arabidopsis. Sci. Rep. 2018, 8, 13349.
- Zhang, S.; Chen, Y.; Zhao, L.; Li, C.; Yu, J.; Li, T.; Yang, W.; Zhang, S.; Su, H.; Wang, L. A novel NAC transcription factor, MdNAC42, regulates anthocyanin accumulation in red-fleshed apple by interacting with MdMYB10. Tree Physiol. 2020, 40, 413–423.
- Wang, R.Z.; Ming, M.L.; Li, J.M.; Shi, D.Q.; Qiao, X.; Li, L.T.; Zhang, S.L.; Wu, J. Genome-wide identification of the MADS-box transcription factor family in pear (Pyrus bretschneideri) reveals evolution and functional divergence. Peerj 2017, 5, e3776.
- Jaakola, L.; Poole, M.; Jones, M.O.; Kamarainen-Karppinen, T.; Koskimaki, J.J.; Hohtola, A.; Haggman, H.; Fraser, P.D.; Manning, K.; King, G.J.; et al. A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol. 2010, 153, 1619–1629.
- Sun, Q.; Jiang, S.; Zhang, T.; Xu, H.; Fang, H.; Zhang, J.; Su, M.; Wang, Y.; Zhang, Z.; Wang, N.; et al. Apple NAC transcription factor MdNAC52 regulates biosynthesis of anthocyanin and proanthocyanidin through MdMYB9 and MdMYB11. Plant Sci. 2019, 289, 110286.
- An, J.P.; Yao, J.F.; Xu, R.R.; You, C.X.; Wang, X.F.; Hao, Y.J. Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation. Plant Cell Environ. 2018, 41, 2678–2692.
- Liu, C.C.; Chi, C.; Jin, L.J.; Zhu, J.; Yu, J.Q.; Zhou, Y.H. The bZip transcription factor HY5 mediates CRY1a-induced anthocyanin biosynthesis in tomato. Plant Cell Environ. 2018, 41, 1762–1775.
- An, J.P.; Qu, F.J.; Yao, J.F.; Wang, X.N.; You, C.X.; Wang, X.F.; Hao, Y.J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 2017, 4, 17023.
- Duan, S.; Wang, J.; Gao, C.; Jin, C.; Li, D.; Peng, D.; Du, G.; Li, Y.; Chen, M. Functional characterization of a heterologously expressed Brassica napus WRKY41-1 transcription factor in regulating anthocyanin biosynthesis in Arabidopsis thaliana. Plant Sci. 2018, 268, 47–53.
- Chen, R.; Cao, Y.; Wang, W.T.; Li, Y.H.; Wang, D.H.; Wang, S.Q.; Cao, X.Y. Transcription factor SmSPL7 promotes anthocyanin accumulation and negatively regulates phenolic acid biosynthesis in Salvia miltiorrhiza. Plant Sci. 2021, 310, 110993.
- Gou, J.Y.; Felippes, F.F.; Liu, C.J.; Weigel, D.; Wang, J.W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 2011, 23, 1512–1522.
- Xie, S.; Lei, Y.J.; Chen, H.W.; Li, J.N.; Chen, H.Z.; Zhang, Z.W. R2R3-MYB Transcription Factors Regulate Anthocyanin Biosynthesis in Grapevine Vegetative Tissues. Front. Plant Sci. 2020, 11, 527.
- Zhong, C.M.; Tang, Y.; Pang, B.; Li, X.K.; Yang, Y.P.; Deng, J.; Feng, C.Y.; Li, L.F.; Ren, G.P.; Wang, Y.Q.; et al. The R2R3-MYB transcription factor GhMYB1a regulates flavonol and anthocyanin accumulation in Gerbera hybrida. Hortic. Res. 2020, 7, 78.
- Shi, S.L.; Liu, Y.; He, Y.J.; Li, L.Z.; Li, D.L.; Chen, H.Y. R2R3-MYB transcription factor SmMYB75 promotes anthocyanin biosynthesis in eggplant (Solanum melongena L.). Sci. Hortic. 2021, 282, 110020.
- Wang, L.J.; Lu, W.X.; Ran, L.Y.; Dou, L.W.; Yao, S.; Hu, J.; Fan, D.; Li, C.F.; Luo, K.M. R2R3-MYB transcription factor MYB6 promotes anthocyanin and proanthocyanidin biosynthesis but inhibits secondary cell wall formation in Populus tomentosa. Plant J. 2019, 99, 733–751.
- Sun, C.; Wang, C.M.; Zhang, W.; Liu, S.A.; Wang, W.Y.; Yu, X.Y.; Song, T.; Yu, M.W.; Yu, W.C.; Qu, S.C. The R2R3-type MYB transcription factor MdMYB90-like is responsible for the enhanced skin color of an apple bud sport mutant. Hortic. Res. 2021, 8, 156.
- Jiang, S.H.; Sun, Q.G.; Zhang, T.L.; Liu, W.J.; Wang, N.; Chen, X.S. MdMYB114 regulates anthocyanin biosynthesis and functions downstream of MdbZIP4-like in apple fruit. J. Plant Physiol. 2021, 257, 153353.
- Zimmermann, I.M.; Heim, M.A.; Weisshaar, B.; Uhrig, J.F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004, 40, 22–34.
- Zuluaga, D.L.; Gonzali, S.; Loreti, E.; Pucciariello, C.; Degl’Innocenti, E.; Guidi, L.; Alpi, A.; Perata, P. Arabidopsis thaliana MYB75/PAP1 transcription factor induces anthocyanin production in transgenic tomato plants. Funct. Plant Biol. 2008, 35, 606–618.
- Jian, W.; Cao, H.H.; Yuan, S.; Liu, Y.D.; Lu, J.F.; Lu, W.; Li, N.; Wang, J.H.; Zou, J.; Tang, N.; et al. SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic. Res. 2019, 6, 22.
- Zhao, M.R.; Li, J.; Zhu, L.; Chang, P.; Li, L.L.; Zhang, L.Y. Identification and Characterization of MYB-bHLH-WD40 Regulatory Complex Members Controlling Anthocyanidin Biosynthesis in Blueberry Fruits Development. Genes 2019, 10, 496.
- Vimolmangkang, S.; Han, Y.P.; Wei, G.C.; Korban, S.S. An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 2013, 13, 176.
- Docimo, T.; Francese, G.; Ruggiero, A.; Batelli, G.; De Palma, M.; Bassolino, L.; Toppino, L.; Rotino, G.L.; Mennella, G.; Tucci, M. Phenylpropanoids Accumulation in Eggplant Fruit: Characterization of Biosynthetic Genes and Regulation by a MYB Transcription Factor. Front. Plant Sci. 2015, 6, 1233.
- Heppel, S.C.; Jaffe, F.W.; Takos, A.M.; Schellmann, S.; Rausch, T.; Walker, A.R.; Bogs, J. Identification of key amino acids for the evolution of promoter target specificity of anthocyanin and proanthocyanidin regulating MYB factors. Plant Mol. Biol. 2013, 82, 457–471.
- Quattrocchio, F.; Wing, J.F.; van der Woude, K.; Mol, J.N.; Koes, R. Analysis of bHLH and MYB domain proteins: Species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. 1998, 13, 475–488.
- Tan, H.; Man, C.; Xie, Y.; Yan, J.; Chu, J.; Huang, J. A Crucial Role of GA-Regulated Flavonol Biosynthesis in Root Growth of Arabidopsis. Mol. Plant 2019, 12, 521–537.
- Zhang, X.; He, Y.; Li, L.; Liu, H.; Hong, G. Involvement of the R2R3-MYB transcription factor MYB21 and its homologs in regulating flavonol accumulation in Arabidopsis stamen. J. Exp. Bot. 2021, 72, 4319–4332.
- Zhi, J.J.; Liu, X.X.; Li, D.J.; Huang, Y.G.M.; Yan, S.S.; Cao, B.H.; Qiu, Z.K. CRISPR/Cas9-mediated SlAN2 mutants reveal various regulatory models of anthocyanin biosynthesis in tomato plant. Plant Cell Rep. 2020, 39, 799–809.
- Sun, C.; Deng, L.; Du, M.; Zhao, J.; Chen, Q.; Huang, T.; Jiang, H.; Li, C.B.; Li, C. A Transcriptional Network Promotes Anthocyanin Biosynthesis in Tomato Flesh. Mol. Plant 2020, 13, 42–58.
- Yan, S.S.; Chen, N.; Huang, Z.J.; Li, D.J.; Zhi, J.J.; Yu, B.W.; Liu, X.X.; Cao, B.H.; Qiu, Z.K. Anthocyanin Fruit encodes an R2R3-MYB transcription factor, SlAN2-like, activating the transcription of SlMYBATV to fine-tune anthocyanin content in tomato fruit. New Phytol. 2020, 225, 2048–2063.
- Li, Y.Q.; Shan, X.T.; Gao, R.F.; Yang, S.; Wang, S.C.; Gao, X.; Wang, L. Two IIIf Clade-bHLHs from Freesia hybrida Play Divergent Roles in Flavonoid Biosynthesis and Trichome Formation when Ectopically Expressed in Arabidopsis. Sci. Rep. 2016, 6, 30514.
- Pires, N.; Dolan, L. Origin and Diversification of Basic-Helix-Loop-Helix Proteins in Plants. Mol. Biol. Evol. 2010, 27, 862–874.
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003, 20, 735–747.
- Baudry, A.; Heim, M.A.; Dubreucq, B.; Caboche, M.; Weisshaar, B.; Lepiniec, L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004, 39, 366–380.
- Zhang, B.P.; Chopra, D.; Schrader, A.; Hulskamp, M. Evolutionary comparison of competitive protein-complex formation of MYB, bHLH, and WDR proteins in plants. J. Exp. Bot. 2019, 70, 3197–3209.
- Spelt, C.; Quattrocchio, F.; Mol, J.N.; Koes, R. anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell. 2000, 12, 1619–1632.
- Fan, Y.; Peng, J.; Wu, J.; Zhou, P.; He, R.; Allan, A.C.; Zeng, L. NtbHLH1, a JAF13-like bHLH, interacts with NtMYB6 to enhance proanthocyanidin accumulation in Chinese Narcissus. BMC Plant Biol. 2021, 21, 275.
- Montefiori, M.; Brendolise, C.; Dare, A.P.; Lin-Wang, K.; Davies, K.M.; Hellens, R.P.; Allan, A.C. In the Solanaceae, a hierarchy of bHLHs confer distinct target specificity to the anthocyanin regulatory complex. J. Exp. Bot. 2015, 66, 1427–1436.
- Wang, L.H.; Tang, W.; Hu, Y.W.; Zhang, Y.B.; Sun, J.Q.; Guo, X.H.; Lu, H.; Yang, Y.; Fang, C.B.; Niu, X.L.; et al. A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant J. 2019, 99, 359–378.




