
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Danny Van Noort | + 1586 word(s) | 1586 | 2021-10-21 08:05:13 | | | |
| 2 | Amina Yu | + 57 word(s) | 1643 | 2021-10-21 10:11:33 | | |
Video Upload Options
Malaria is caused by a protozoan parasite called Plasmodium. Four different species of Plasmodium cause human infections: P. falciparum, P. malariae, P. ovale and P. vivax. Worldwide, the most prevalent species is P. falciparum (around 96%), which is the most lethal species (300,000 deaths worldwide in 2017), and P. vivax, which has the most extensive geographical distribution.
1. Introduction
In contrast with other pathogens, Plasmodium efficiently infects the host by entering cells. In mammalian hosts, they are transmitted by the bite of the female Anopheles mosquito infected with Plasmodium spp. , which it houses in its salivary glands as sporozoites. When a mosquito feeds, the sporozites are injected into the hosts’ bloodstream [1][2][3]. Another possible source of infection is blood transfusions.
In humans, the parasite has two cycles: exoerythrocytic and erythrocytic ( Figure 1 ). During the exoerythrocytic cycle, Plasmodium spp. matures to the hypnozoite form and infects hepatocytes to become hepatic schizonts; these can remain in the liver for many years [4]. The parasite breaks through hepatocytes at the merozoite stage to infect red blood cells (RBC). It develops into early trophozoites (ring form), late trophozoites, and finally into erythrocytic schizonts. They can also turn into gametocytes and can then be suctioned by another Anopheles spp., thus repeating the cycle [5]. Plasmodium adheres to the RBC via a high-affinity ligand–receptor interaction from where it will invade the cell. An infection will cause extensive changes to the RBC, such as a change in cell morphology, increased rigidity, elevated permeability, and changes in iron content [6]. These changes in the RBCs can lead to cerebral malaria or anemia.
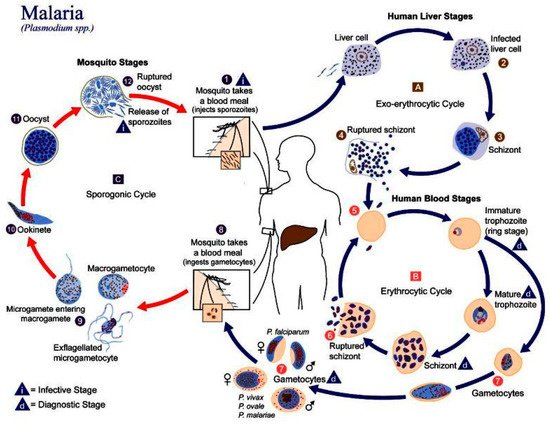
2. Malaria in Peru
Malaria has been a health issue in Peru since colonial times and is thought to have arrived with the migration of slaves from Africa. In the 17th century, the discovery of quinine from the endemic tree of Peru Cinchona officinalis was decisive for treating patients with this disease. Ever since, malaria has been a primary national health concern in Peru [7][8].
Epidemiologically, Peru follows the trend of the American Region, with P. vivax being the predominant species in the country (80.2% of the total cases), followed by P. falciparum (19.8%); the provinces with the most cases are Loreto, Amazonas, and Junin (95.8% of cases in the country) [9]. The lack of access to health centers and the fact that malaria shares some symptoms with other endemic diseases make the diagnosis more difficult and time-demanding, enabling the progression of the disease. It is calculated that the treatment of malaria costs a third of the family income of each patient [10][11]. To control the disease, different administrations have created policies to address different aspects of the problem: epidemiological control, prevention programs, provision of different medicines according to epidemiological characteristics, the control of Anopheles spp. , and the application and design of different screening methods [8]. Since 2018, Peru has incorporated the Malaria Zero Program, a community-focused approach, and attempts to eradicate the disease by 2030. Unfortunately, with the outbreak of COVID-19, the activities of this program have been severely affected, primarily epidemiological surveillance, patient treatment, research projects, and educational activities in communities [11].
The most used diagnostic method in Peru is the Giemsa stain. Although its precision and low cost make it a powerful method, its reliance on infrastructure and specialists makes diagnostics with this technique challenging in poor and remote areas [10]. Another popular diagnostic method is PCR; its popularity is due to its high precision and throughput. However, the cost of PCR tests makes them prohibitively expensive for most of the country. That is why we, as a research group in Peru, are looking for a cheaper and faster way to perform diagnostics.
3. Current Methods of Diagnostics
Fast diagnosis and prompt treatment are necessary to prevent mild cases of Malaria from turning more serious, which could result in death. Presently, most diagnoses are performed with either optical means, which requires a microscope, or so-called rapid diagnostic tests (RDTs). The most commonly used optical method is the blood smear, which determines the presence of malaria parasites in the blood cells [12]. However, a low concentration of the malaria parasite can result in false negatives, as there might not be a parasite in the sample taken and imaged under the microscope [13]. The detection threshold under field conditions is estimated to be 50–100 parasites/mL [14][15]. In a laboratory setting, this threshold is 4–20 parasites/mL [16]. There can also be errors in species identification. Failures to differentiate P. falciparum from P. vivax , the two most common species of malaria, are frequent in routine microscopic diagnostics, and are therefore under-reported [15].
The same can be said for RDTs. RDTs rely on antigen–antibody interaction, resulting in a visible band on a paper strip, in the same manner as pregnancy tests. However, if there is a low concentration of malaria parasites in the blood, no band will show, leading to a negative result. The other drawback is that RDT does not screen for multiple strains of the parasite. Another promising POC method to detect malaria is loop-mediated isothermal amplification (LAMP). This method is able to detect quantities of the parasite that are below the detection threshold for other methods. LAMP has been shown to achieve specificities of up to 100% and sensitivities up to 98% [17].
Other methods, such as DNA probes and PCR [18], fluorescent staining using flow cytometry [19], detection of malaria pigments by depolarized laser light [20], and mass spectrometry [21], are either not suitable for point-of-care (POCs) in remote locations or show limited success.
Therefore, better, i.e., more sensitive, differential, and faster, POC diagnostics methods are required, which can be used in remote locations when necessary. Microfluidics can play an important role in such devices. This review will give an overview of existing microfluidic-based devices that can be used for either the detection, separation, or diagnosis. At the end of the review, we will also suggest other devices envisioned to help combat malaria on all levels.
4. Paper-Based Microfluidics and Malaria
One method that is suitable for use in remote locations because of its low cost and ease of operation is paper-based microfluidics, otherwise known as lateral flow devices [21]. Because of the properties of paper, these devices use capillary forces to pump fluids laterally through the paper, an example of this can be seen in Figure 2 . Here, a paper lateral flow device (item 2) is used to perform a multistep assay [22]. Advances in paper-based microfluidic analysis have addressed some of the limitations previously associated with lateral flow devices, such as the inability to create valves [23] and multiplex or perform multi-step analysis [24]. Paper-based lateral flow devices have been primarily used to automate multistep assays quickly and cheaply. For malaria detection, the literature shows that loop-mediated isothermal amplification (LAMP) and signal-amplified sandwich format immunoassay for PfHRP2 are popular methods of detecting the presence of an infection. The results from these methods have been shown to be comparable to those obtained using ELISA [25].
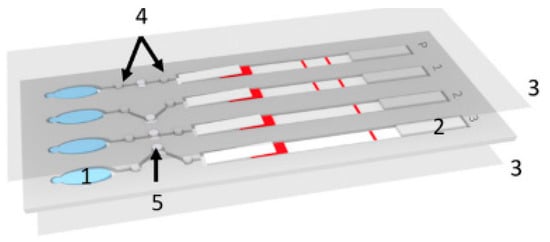
Implementation of improved signal amplification techniques has increased the reliability of the readout [26]. Some of these improvements have been applied to detecting malaria in whole and pretreated blood, such as the detection of specific proteins in a blood sample. Fu et al. used a 2D paper network to filter whole blood and deliver appropriate volumes of the sample mixed with antibodies, rinse buffer, and a signal amplification reagent [25]. The device consists of a horizontal channel with a large pad (reaction area) at the end. The channel has three side channels at different positions down the main channel. The distance between the side channels and the reaction area determines the time it takes for the reagent to flow through the side channel to reach the reaction pad. With this device, Fu et al. were able to identify fetal bovine serum (FBS) spiked with different amounts of Plasmodium falciparum histidine rich protein PfHRP2. They showed that this technique is very promising as it is easy to use and can store the reagents on the device in a dehydrated form.
Another method to control the timing of reactions in a paper microfluidic device is by using microfluidic actuators (sponges), which expand at a known rate [27]. Toley et al. used this method to perform a similar process as Hu et al. and successfully detected PfHRP2 in a sample.
Other paper-microfluidic devices can process pretreated blood or preconcentrate samples using magnetic markers. Using a simple 1D paper microfluidic device, researchers performed DNA extraction and loop-mediated isothermal amplification (LAMP), and detected DNA amplicons using lateral flow and antibody-based detection [26]. This device has a section that multiplexes the LAMP amplification and performs it three times as well as a control test. First, the sample was pretreated with functionalized magnetic particles which bound to Plasmodium falciparum L-lactate dehydrogenase (PfLDH). The sample was then loaded onto the chip, and a magnetic lens was placed underneath. By applying a current to the lens, the researchers formed virtual channels that connected the sample input to an absorbent wick. The magnetically tagged analytes were then trapped in the magnetic field while the supernatant was absorbed, leaving a large concentration of tagged PfLDH [26]. Paper microfluidics have also been shown to be useful in simpler preconcentration methods. One such application was shown by Bahmla et al. who used a circular paper disc, capillary tubes, and inertial forces to turn the disc into a low-cost centrifuge [28].
References
- Cowman, A.F.; Crabb, B.S. Invasion of red blood cells by malaria parasites. Cell 2006, 124, 755–766.
- Baia-da-Silva, D.C.; Alvarez, L.C.S.; Lizcano, O.V.; Costa, F.T.M.; Lopes, S.C.P.; Orfanó, A.S.; Pascoal, D.O.; Nacif-Pimenta, R.; Rodriguez, I.C.; Guerra, M.d.G.V.B. The role of the peritrophic matrix and red blood cell con-centration in Plasmodium vivax infection of Anopheles aquasalis. Parasites Vectors 2018, 11, 1–10.
- Choumet, V.; Carmi-Leroy, A.; Laurent, C.; Lenormand, P.; Rousselle, J.-C.; Namane, A.; Roth, C.; Brey, P.T. The salivary glands and saliva of Anopheles gambiae as an essential step in the Plasmodium life cycle: A global proteomic study. Proteomics 2007, 7, 3384–3394.
- Schafer, C.; Dambrauskas, N.; Steel, R.; Carbonetti, S.; Chuenchob, V.; Flannery, E.L.; Vigdorovich, V.; Oliver, B.; Roobsoong, W.; Maher, S.P.; et al. A recombinant antibody against Plasmodium vivax UIS4 for distinguishing replicating from dormant liver stages. Malaria J. 2018, 17, 1–10.
- Beck, T. Will malaria soon be a thing of the past? In Coffee Break: Tutorials for NCBI Tools ; National Center for Biotechnology Information: Bethesda, MD, USA, 2006.
- Mohandas, N.; An, X. Malaria and human red blood cells. Med. Microbiol. Immunol. 2012, 201, 593–598.
- Legua, P. Malaria en el Perú. Rev. Med. Herediana 1994, 5, 115–117.
- Herrera, J.V. Prevención y control de la Malaria y otras enfermedades trasmitidas por vectores en el Perú. Rev. Peru. Epidemiol. 2003, 11, 18.
- Boletin Epidemiologico Se 48 Minsa; Ministerio de Salud: Lima, Peru, 2020.
- Cabezas, C. Pruebas rápidas para el diagnóstico de la malaria: Una necesidad en áreas rurales con limitado acceso al diagnóstico microscópico. Rev. Peru. Med. Exp. Salud Pública 2006, 23, 79–80.
- Torres, K.; Alava, F.; Soto-Calle, V.; Llanos-Cuentas, A.; Rodriguez, H.; Llacsahuanga, L.; Gamboa, D.; Vinetz, J. Malaria situation in the Peruvian Amazon during the COVID-19 pandemic. Am. J. Trop. Med. Hyg. 2020, 103, 1773–1776.
- Halim, S.; Bretschneider, T.R.; Li, Y.; Preiser, P.R.; Kuss, C. Estimating malaria parasitaemia from blood smear image. In Proceedings of the 9th International Conference on Control, Automation, Robotics and Vision (ICARCV 2006), Singapore, 5–8 December 2006; pp. 1–6.
- Maguire, J.D.; Lederman, E.R.; Barcus, M.J.; O’Meara, W.A.P.; Jordon, R.G.; Duong, S.; Muth, S.; Sismadi, P.; Bangs, M.J.; Prescott, W.R.; et al. Production and validation of durable, high quality standardized malaria microscopy slides for teaching, testing and quality assurance during an era of declining diagnostic proficiency. Malaria J. 2006, 5, 92.
- World Health Organization. Malaria diagnosis: Memorandum from a WHO meeting. Bull. World Health Organ. 1988, 66, 575–594.
- Milne, L.; Kyi, M.; Chiodini, P.; Warhurst, D. Accuracy of routine laboratory diagnosis of malaria in the United Kingdom. J. Clin. Pathol. 1994, 47, 740–742.
- Payne, D. Use and limitations of light microscopy for diagnosing malaria at the primary health care level. Bull. World Health Organ. 1988, 66, 621.
- Hayashida, K.; Kajino, K.; Simukoko, H.; Simuunza, M.; Ndebe, J.; Chota, A.; Namangala, B.; Sugimoto, C. Direct detection of falciparum and non-falciparum malaria DNA from a drop of blood with high sensitivity by the dried-LAMP system. Parasites Vectors 2017, 10, 1–9.
- Snounou, G.; Viriyakosol, S.; Jarra, W.; Thaithong, S.; Brown, K. Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol. Biochem. Parasitol. 1993, 58, 283–292.
- van Vianen, P.H.; van Engen, A.; Thaithong, S.; van der Keur, M.; Tanke, H.J.; van der Kaay, H.J.; Mons, B.; Janse, C.J. Flow cytometric screening of blood samples for malaria parasites. Cytometry 1993, 14, 276–280.
- Mendelow, B.V.; Lyons, C.; Nhlangothi, P.; Tana, M.; Munster, M.; Wypkema, E.; Liebowitz, L.; Marshall, L.; Scott, S.; Coetzer, T.L. Automated malaria detection by depolarization of laser light. Br. J. Haematol. 1999, 104, 499–503.
- Wongsrichanalai, C.; Baracus, M.J.; Muth, S.; Sutamihardja, A.; Wernsdorfer, W.H. A review of malaria diagnostic tools: Microscopy and rapid diagnostic test (RDT). Am. J. Trop. Med. Hyg. 2007, 77 (Suppl. 6), 119–127.
- Reboud, J.; Xu, G.; Garrett, A.; Adriko, M.; Yang, Z.; Tukahebwa, E.M.; Rowell, C.; Cooper, J.M. Paper based microfluidics for DNA diagnostics of malaria in low resource underserved rural communities. Proc. Natl. Acad. Sci. USA 2019, 116, 4834–4842.
- Kim, T.H.; Hahn, Y.K.; Lee, J.; van Noort, D.; Kim, M.S. Solenoid driven pressure valve system: Toward versatile fluidic control in paper microfluidics. Anal. Chem. 2018, 90, 2534–2541.
- Xu, G.; Nolder, D.; Reboud, J.; Oguike, M.C.; van Schalkwyk, D.; Sutherland, C.J.; Cooper, J.M. Paper-origami-based multiplexed malaria diagnostics from whole blood. Angew. Chem. 2016, 128, 15476–15479.
- Fu, E.; Liang, T.; Spicar-Mihalic, P.; Houghtaling, J.; Ramachandran, S.; Yager, P. Two-Dimensional Paper Network Format That Enables Simple Multistep Assays for Use in Low-Resource Settings in the Context of Malaria Antigen Detection. Anal. Chem. 2012, 84, 4574–4579.
- Rackus, D.G.; de Campos, R.P.S.; Chan, C.; Karcz, M.M.; Seale, B.; Narahari, T.; Dixon, C.; Chamberlainab, M.D.; Wheeler, A.R. Pre-concentration by liquid intake by paper (P-CLIP): A new technique for large volumes and digital microfluidics. Lab Chip 2017, 17, 2272–2280.
- Toley, B.J.; Wang, J.A.; Gupta, M.; Buser, J.R.; LaFleur, L.K.; Lutz, B.R.; Fu, E.; Yager, P. A versatile valving toolkit for automating fluidic operations in paper microfluidic devices. Lab Chip 2015, 15, 1432–1444.
- Bhamla, M.S.; Benson, B.; Chai, C.; Katsikis, G.; Johri, A.; Prakash, M. Hand-powered ultralow-cost paper centrifuge. Nat. Biomed. Eng. 2017, 1, 1–7.




