
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Noman Shoaib | + 3342 word(s) | 3342 | 2021-10-12 05:46:50 | | | |
| 2 | Peter Tang | Meta information modification | 3342 | 2021-10-14 04:48:44 | | |
Video Upload Options
Starch phosphorylase is a member of the GT35-glycogen-phosphorylase superfamily. Glycogen phosphorylases have been researched in animals thoroughly when compared to plants. Genetic evidence signifies the integral role of plastidial starch phosphorylase (PHO1) in starch biosynthesis in model plants. The counterpart of PHO1 is PHO2, which specifically resides in cytosol and is reported to lack L80 peptide in the middle region of proteins as seen in animal and maltodextrin forms of phosphorylases. The function of this extra peptide varies among species and ranges from the substrate of proteasomes to modulate the degradation of PHO1 in Solanum tuberosum to a non-significant effect on biochemical activity in Oryza sativa and Hordeum vulgare.
1. Introduction
2. Starch Metabolism and Phosphorylase
2.1. Starch Biosynthesis
2.2. Initiation of Starch Granule in Comparison to Glycogen
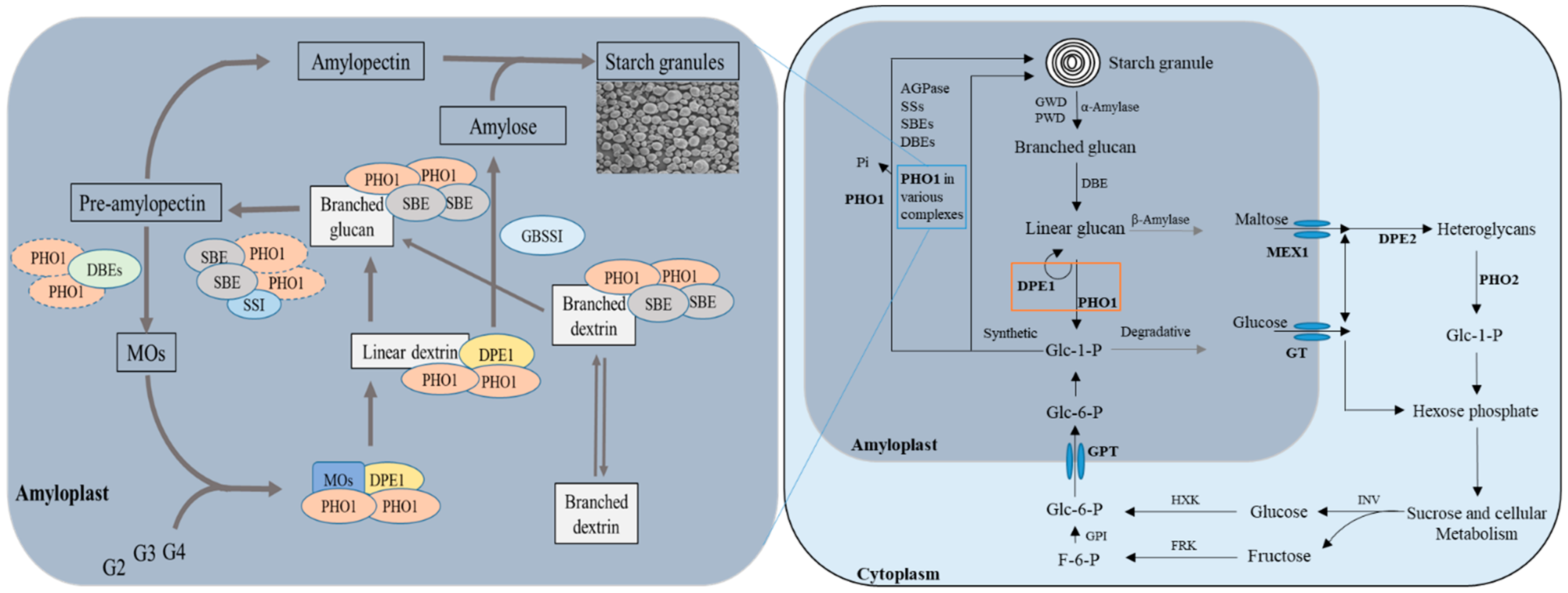
2.3. Multiple Complex Formation
2.4. Starch Degradation
2.5. Additional Role of PHO1
3. Synthetic and Phosphorolytic Activity of Phosphorylase
4. Genomics of PHO1
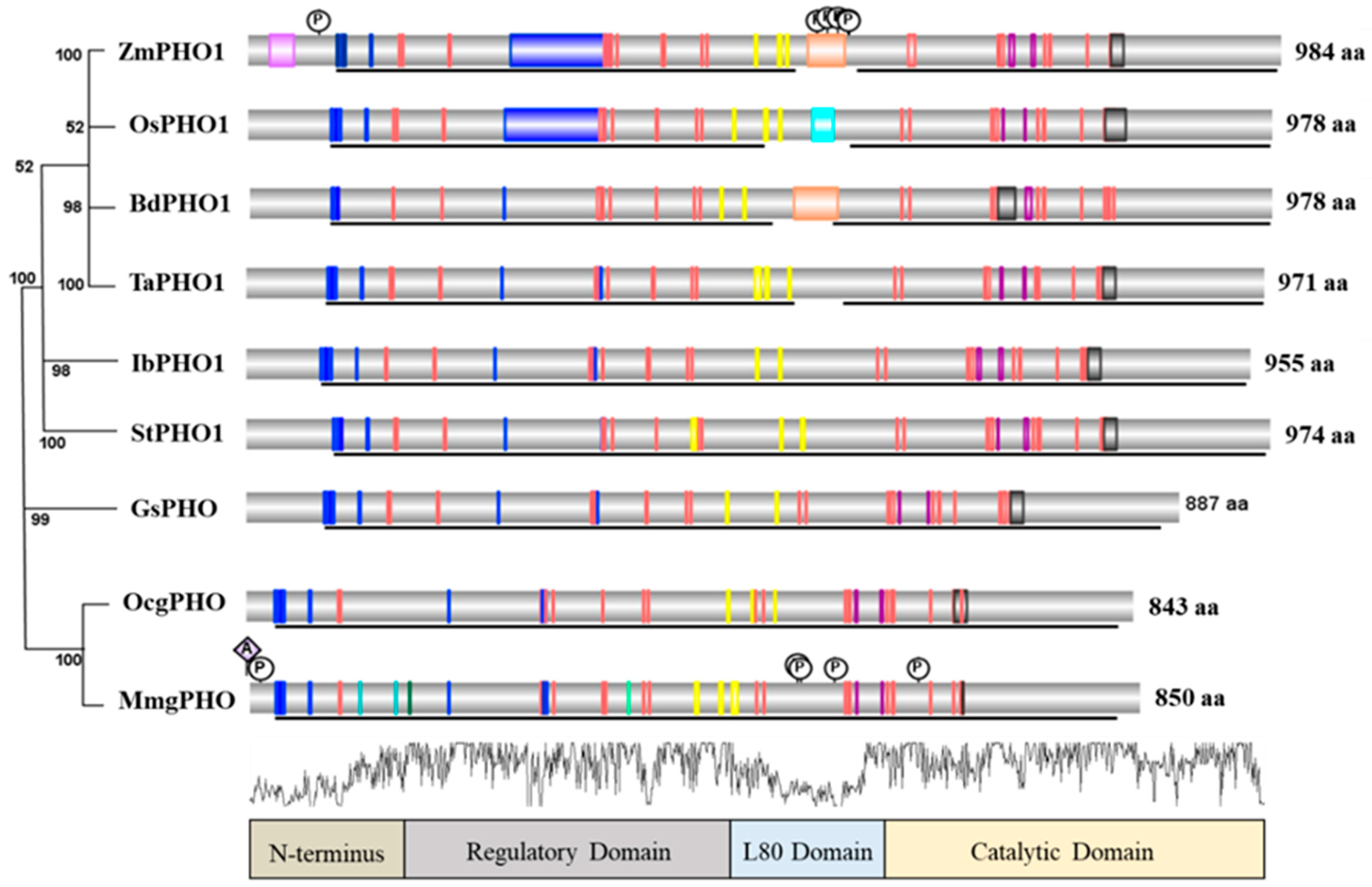
5. Structure and Comparison of PHO1

References
- Cori, G.T.; Cori, C.F.; Schmidt, G. The role of glucose-l-phosphate in the formation of blood sugar and synthesis of glycogen in the liver. J. Biol. Chem. 1939, 142, 355–366.
- Green, D.E.S.; Stumpf, P.K. Starch Phosphorylase of potato. J. Biol. Chem. 1942, 142, 355–366.
- Hanes, C.S. The breakdown and synthesis of starch by an enzyme system from pea seeds. Proc. R. Soc. Lond. Ser. B Boil. Sci. 1940, 128, 421–450.
- Tsai, C.Y.; Nelson, O.E. Phosphorylases I and II of Maize Endosperm. Plant Physiol. 1968, 43, 103–112.
- Subasinghe, R.M.; Liu, F.; Polack, U.C.; Lee, E.A.; Emes, M.J.; Tetlow, I.J. Multimeric states of starch phosphorylase determine protein–protein interactions with starch biosynthetic enzymes in amyloplasts. Plant Physiol. Biochem. 2014, 83, 168–179.
- Nakano, K.; Fukui, T. The complete amino acid sequence of potato alpha-glucan phosphorylase. J. Biol. Chem. 1986, 261, 8230–8236.
- Brisson, N.; Giroux, H.; Zollinger, M.; Camirand, A.; Simard, C. Maturation and Subcellular Compartmentation of Potato Starch Phosphorylase. Plant Cell 1989, 1, 559.
- Yu, Y.; Mu, H.H.; Wasserman, B.P.; Carman, G.M. Identification of the Maize Amyloplast Stromal 112-kD Protein as a Plastidic Starch Phosphorylase. Plant Physiol. 2001, 125, 351–359.
- Tickle, P.; Burrell, M.M.; Coates, S.A.; Emes, M.J.; Tetlow, I.J.; Bowsher, C.G. Characterization of plastidial starch phosphorylase in Triticum aestivum L. endosperm. J. Plant Physiol. 2009, 166, 1465–1478.
- Hwang, S.-K.; Singh, S.; Çakir, B.; Satoh, H.; Okita, T.W. The plastidial starch phosphorylase from rice endosperm: Catalytic properties at low temperature. Planta 2016, 243, 999–1009.
- Cuesta-Seijo, J.A.; Nielsen, M.M.; Ruzanski, C.; Krucewicz, K.; Beeren, S.R.; Rydhal, M.G.; Yoshimura, Y.; Striebeck, A.; Motawia, M.S.; Willats, W.G.T.; et al. In Vitro Biochemical Characterization of All Barley Endosperm Starch Synthases. Front. Plant Sci. 2016, 6, 1265.
- Hwang, P.; Koper, K.; Okita, T.W. The plastid phosphorylase as a multiple-role player in plant metabolism. Plant Sci. 2020, 290, 110303.
- Albrecht, T.; Greve, B.; Pusch, K.; Kossmann, J.; Buchner, P.; Wobus, U.; Steup, M. Homodimers and heterodimers of Pho1-type phosphorylase isoforms in Solanum tuberosum L. as revealed by sequence-specific antibodies. JBIC J. Biol. Inorg. Chem. 1998, 251, 343–352.
- Lin, Y.-C.; Chen, H.-M.; Chou, I.-M.; Chen, A.-N.; Chen, C.-P.; Young, G.-H.; Lin, C.-T.; Cheng, C.-H.; Chang, S.-C.; Juang, R.-H. Plastidial Starch Phosphorylase in Sweet Potato Roots Is Proteolytically Modified by Protein-Protein Interaction with the 20S Proteasome. PLoS ONE 2012, 7, e35336.
- Ko, Y.-T.; Chang, J.-Y.; Lee, Y.-T.; Wu, Y.-H. The Identification of Starch Phosphorylase in the Developing Mungbean (Vigna radiata L.). J. Agric. Food Chem. 2005, 53, 5708–5715.
- Satoh, H.; Shibahara, K.; Tokunaga, T.; Nishi, A.; Tasaki, M.; Hwang, S.-K.; Okita, T.; Kaneko, N.; Fujita, N.; Yoshida, M.; et al. Mutation of the Plastidial α-Glucan Phosphorylase Gene in Rice Affects the Synthesis and Structure of Starch in the Endosperm. Plant Cell 2008, 20, 1833–1849.
- Hwang, S.-K.; Koper, K.; Satoh, H.; Okita, T.W. Rice Endosperm Starch Phosphorylase (Pho1) Assembles with Disproportionating Enzyme (Dpe1) to Form a Protein Complex That Enhances Synthesis of Malto-oligosaccharides. J. Biol. Chem. 2016, 291, 19994–20007.
- Cuesta-Seijo, J.A.; Ruzanski, C.; Krucewicz, K.; Meier, S.; Hägglund, P.; Svensson, B.; Palcic, M.M. Functional and structural characterization of plastidic starch phosphorylase during barley endosperm development. PLoS ONE 2017, 12, e0175488.
- Maddelein, M.L.; Libessart, N.; Bellanger, F.; Delrue, B.; D’Hulst, C.; Koornhuyse, N.V.D.; Fontaine, T.; Wieruszeski, J.M.; Decq, A.; Ball, S. Toward an understanding of the biogenesis of the starch granule. Determination of granule-bound and soluble starch synthase functions in amylopectin synthesis. J. Biol. Chem. 1994, 269, 25150–25157.
- Putseys, J.A.; Derde, L.J.; Lamberts, L.; Östman, E.; Björck, I.M.; Delcour, J.A. Functionality of Short Chain Amylose-Lipid Complexes in Starch-Water Systems and Their Impact on In Vitro Starch Degradation. J. Agric. Food Chem. 2010, 58, 1939–1945.
- Goren, A.; Ashlock, D.; Tetlow, I.J. Starch formation inside plastids of higher plants. Protoplasma 2018, 255, 1855–1876.
- Yu, G.; Lv, Y.; Shen, L.; Wang, Y.; Qing, Y.; Wu, N.; Li, Y.; Huang, H.; Zhang, N.; Liu, Y.; et al. The Proteomic Analysis of Maize Endosperm Protein Enriched by Phos-tag(tm) Reveals the Phosphorylation of Brittle-2 Subunit of ADP-Glc Pyrophosphorylase in Starch Biosynthesis Process. Int. J. Mol. Sci. 2019, 20, 986.
- Fujita, N.; Kubo, A.; Suh, D.-S.; Wong, K.-S.; Jane, J.-L.; Ozawa, K.; Takaiwa, F.; Inaba, Y.; Nakamura, Y. Antisense inhibition of isoamylase alters the structure of amylopectin and the physicochemical properties of starch in rice endosperm. Plant Cell Physiol. 2003, 44, 607–618.
- Fujita, N.; Yoshida, M.; Asakura, N.; Ohdan, T.; Miyao, A.; Hirochika, H.; Nakamura, Y. Function and Characterization of Starch Synthase I Using Mutants in Rice. Plant Physiol. 2006, 140, 1070–1084.
- Hanashiro, I.; Itoh, K.; Kuratomi, Y.; Yamazaki, M.; Igarashi, T.; Matsugasako, J.-I.; Takeda, Y. Granule-Bound Starch Synthase I is Responsible for Biosynthesis of Extra-Long Unit Chains of Amylopectin in Rice. Plant Cell Physiol. 2008, 49, 925–933.
- Ortiz-Marchena, M.I.; Albi, T.; Lucas-Reina, E.; Said, F.E.; Romero-Campero, F.J.; Cano, B.; Ruiz, M.T.; Romero, J.M.; Valverde, F. Photoperiodic Control of Carbon Distribution during the Floral Transition in Arabidopsis. Plant Cell 2014, 26, 565–584.
- Streb, S.; Delatte, T.; Umhang, M.; Eicke, S.; Schorderet, M.; Reinhardt, D.; Zeeman, S.C. Starch Granule Biosynthesis in Arabidopsis Is Abolished by Removal of All Debranching Enzymes but Restored by the Subsequent Removal of an Endoamylase. Plant Cell 2009, 20, 3448–3466.
- Pfister, B.; Zeeman, S.C. Formation of starch in plant cells. Cell. Mol. Life Sci. 2016, 73, 2781–2807.
- Delatte, T.; Umhang, M.; Trevisan, M.; Eicke, S.; Thorneycroft, D.; Smith, S.; Zeeman, S. Evidence for Distinct Mechanisms of Starch Granule Breakdown in Plants. J. Biol. Chem. 2006, 281, 12050–12059.
- Ma, J.; Jiang, Q.-T.; Zhang, X.-W.; Lan, X.-J.; Pu, Z.-E.; Wei, Y.-M.; Liu, C.; Lu, Z.-X.; Zheng, Y.-L. Structure and expression of barley starch phosphorylase genes. Planta 2013, 238, 1081–1093.
- Higgins, J.E.; Kosar-Hashemi, B.; Li, Z.; Howitt, C.A.; Larroque, O.; Flanagan, B.; Morell, M.K.; Rahman, S. Characterization of starch phosphorylases in barley grains. J. Sci. Food Agric. 2013, 93, 2137–2145.
- Zeeman, S.C.; Thorneycroft, D.; Schupp, N.; Chapple, A.; Weck, M.; Dunstan, H.; Haldimann, P.; Bechtold, N.; Smith, A.M.; Smith, S. Plastidial α-Glucan Phosphorylase Is Not Required for Starch Degradation in Arabidopsis Leaves but Has a Role in the Tolerance of Abiotic Stress. Plant Physiol. 2004, 135, 849–858.
- Tetlow, I.J.; Bertoft, E. A Review of Starch Biosynthesis in Relation to the Building Block-Backbone Model. Int. J. Mol. Sci. 2020, 21, 7011.
- Roldán, I.; Wattebled, F.; Lucas, M.M.; Delvallé, D.; Planchot, V.; Jiménez, S.; Pérez, R.; Ball, S.; D’Hulst, C.; Mérida, Á. The phenotype of soluble starch synthase IV defective mutants of Arabidopsis thaliana suggests a novel function of elongation enzymes in the control of starch granule formation. Plant J. 2007, 49, 492–504.
- Leterrier, M.; Holappa, L.D.; Broglie, K.E.; Beckles, D.M. Cloning, characterisation and comparative analysis of a starch synthase IV gene in wheat: Functional and evolutionary implications. BMC Plant Biol. 2008, 8, 98.
- Roach, P.J.; DePaoli-Roach, A.A.; Hurley, T.; Tagliabracci, V.S. Glycogen and its metabolism: Some new developments and old themes. Biochem. J. 2012, 441, 763–787.
- Burrell, M.M. Starch: The need for improved quality or quantity—An overview. J. Exp. Bot. 2003, 54, 451–456.
- Tetlow, I.J.; Emes, M.J. A review of starch-branching enzymes and their role in amylopectin biosynthesis. IUBMB Life 2014, 66, 546–558.
- Crofts, N.; Nakamura, Y.; Fujita, N. Critical and speculative review of the roles of multi-protein complexes in starch biosynthesis in cereals. Plant Sci. 2017, 262, 1–8.
- Illingworth, B.; Brown, D.H.; Cori, C.F. The de novo synthesis of polysaccharide by phosphorylase. Proc. Natl. Acad. Sci. USA 1961, 47, 469–478.
- Tsai, C.Y.; Nelson, O.E. Two Additional Phosphorylases in Developing Maize Seeds. Plant Physiol. 1969, 44, 159–167.
- Grimaud, F.; Rogniaux, H.; James, M.G.; Myers, A.; Planchot, V. Proteome and phosphoproteome analysis of starch granule-associated proteins from normal maize and mutants affected in starch biosynthesis. J. Exp. Bot. 2008, 59, 3395–3406.
- Tetlow, I.J.; Beisel, K.G.; Cameron, S.; Makhmoudova, A.; Liu, F.; Bresolin, N.S.; Wait, R.; Morell, M.K.; Emes, M.J. Analysis of Protein Complexes in Wheat Amyloplasts Reveals Functional Interactions among Starch Biosynthetic Enzymes. Plant Physiol. 2008, 146, 1878–1891.
- Hwang, S.-K.; Nishi, A.; Satoh, H.; Okita, T. Rice endosperm-specific plastidial α-glucan phosphorylase is important for synthesis of short-chain malto-oligosaccharides. Arch. Biochem. Biophys. 2010, 495, 82–92.
- Ozbun, J.L.; Hawker, J.S.; Greenberg, E.; Lammel, C.; Preiss, J.; Lee, E.Y.C. Starch Synthetase, Phosphorylase, ADPglucose Pyrophosphorylase, and UDPglucose Pyrophosphorylase in Developing Maize Kernels. Plant Physiol. 1973, 51, 1–5.
- Dauvillée, D.; Chochois, V.; Steup, M.; Haebel, S.; Eckermann, N.; Ritte, G.; Ral, J.-P.; Colleoni, C.; Hicks, G.; Wattebled, F.; et al. Plastidial phosphorylase is required for normal starch synthesis in Chlamydomonas reinhardtii. Plant J. 2006, 48, 274–285.
- Nakamura, Y.; Ono, M.; Sawada, T.; Crofts, N.; Fujita, N.; Steup, M. Characterization of the functional interactions of plastidial starch phosphorylase and starch branching enzymes from rice endosperm during reserve starch biosynthesis. Plant Sci. 2017, 264, 83–95.
- Rathore, R.; Garg, N.; Garg, S.; Kumar, A. Starch phosphorylase: Role in starch metabolism and biotechnological applications. Crit. Rev. Biotechnol. 2009, 29, 214–224.
- Nakamura, Y.; Ono, M.; Utsumi, C.; Steup, M. Functional Interaction Between Plastidial Starch Phosphorylase and Starch Branching Enzymes from Rice During the Synthesis of Branched Maltodextrins. Plant Cell Physiol. 2012, 53, 869–878.
- Crofts, N.; Abe, N.; Oitome, N.F.; Matsushima, R.; Hayashi, M.; Tetlow, I.J.; Emes, M.J.; Nakamura, Y.; Fujita, N. Amylopectin biosynthetic enzymes from developing rice seed form enzymatically active protein complexes. J. Exp. Bot. 2015, 66, 4469–4482.
- Colleoni, C.; Dauvillée, D.; Mouille, G.; Morell, M.; Samuel, M.; Slomiany, M.-C.; Liénard, L.; Wattebled, F.; D’Hulst, C.; Ball, S. Biochemical Characterization of the Chlamydomonas reinhardtii α-1,4 Glucanotransferase Supports a Direct Function in Amylopectin Biosynthesis. Plant Physiol. 1999, 120, 1005–1014.
- Zeeman, S.C.; Delatte, T.; Messerli, G.; Umhang, M.; Stettler, M.; Mettler, T.; Streb, S.; Reinhold, H.; Kötting, O. Starch breakdown: Recent discoveries suggest distinct pathways and novel mechanisms. Funct. Plant Biol. 2007, 34, 465–473.
- Orzechowski, S. Starch metabolism in leaves. Acta Biochim. Pol. 2008, 55, 435–445.
- Fettke, J.; Eckermann, N.; Kötting, O.; Ritte, G.; Steup, M. Novel starch-related enzymes and carbohydrates. Cell. Mol. Boil. 2007, 52, 883–904.
- Mu, H.H.; Yu, Y.; Wasserman, B.P.; Carman, G.M. Purification and Characterization of the Maize Amyloplast Stromal 112-kDa Starch Phosphorylase. Arch. Biochem. Biophys. 2001, 388, 155–164.
- Mizuno, S.; Kamiyoshihara, Y.; Shiba, H.; Shinmachi, F.; Watanabe, K.; Tateishi, A. Plastidial starch phosphorylase is highly associated with starch accumulation process in developing squash (Cucurbita sp.) fruit. Physiol. Plant. 2018, 167, 264–275.
- Tiessen, A.; Nerlich, A.; Faix, B.; Hümmer, C.; Fox, S.; Trafford, K.; Weber, H.; Weschke, W.; Geigenberger, P. Subcellular analysis of starch metabolism in developing barley seeds using a non-aqueous fractionation method. J. Exp. Bot. 2012, 63, 2071–2087.
- Fettke, J.; Albrecht, T.; Hejazi, M.; Mahlow, S.; Nakamura, Y.; Steup, M. Glucose 1-phosphate is efficiently taken up by potato (Solanum tuberosum) tuber parenchyma cells and converted to reserve starch granules. N. Phytol. 2010, 185, 663–675.
- Malinova, I.; Mahlow, S.; Alseekh, S.; Orawetz, T.; Fernie, A.R.; Baumann, O.; Steup, M.; Fettke, J. Double Knockout Mutants of Arabidopsis Grown under Normal Conditions Reveal that the Plastidial Phosphorylase Isozyme Participates in Transitory Starch Metabolism. Plant Physiol. 2014, 164, 907–921.
- Jiang, J.; Yao, C.; Cao, X.; Liu, Y.; Xue, S. Characterization of starch phosphorylase from the marine green microalga (Chlorophyta) Tetraselmis subcordiformis reveals its potential role in starch biosynthesis. J. Plant Physiol. 2017, 218, 84–93.
- Fettke, J.; Leifels, L.; Brust, H.; Herbst, K.; Steup, M. Two carbon fluxes to reserve starch in potato (Solanum tuberosum L.) tuber cells are closely interconnected but differently modulated by temperature. J. Exp. Bot. 2012, 63, 3011–3029.
- Bustos, R.; Fahy, B.; Hylton, C.M.; Seale, R.; Nebane, N.M.; Edwards, A.; Martin, C.; Smith, A.M. Starch granule initiation is controlled by a heteromultimeric isoamylase in potato tubers. Proc. Natl. Acad. Sci. USA 2004, 101, 2215–2220.
- Tetlow, I.J.; Wait, R.; Lu, Z.; Akkasaeng, R.; Bowsher, C.G.; Esposito, S.; Kosar-Hashemi, B.; Morell, M.K.; Emes, M.J. Protein Phosphorylation in Amyloplasts Regulates Starch Branching Enzyme Activity and Protein–Protein Interactions. Plant Cell 2004, 16, 694–708.
- Subasinghe, R.M. Role and Regulation of Starch Phosphorylase and Starch Synthase IV in Starch Biosynthesis in Maize Endosperm Amyloplast. Ph.D. Thesis, University of Guelph, Guelph, ON, Canada, 2014.
- Young, G.-H.; Chen, H.-M.; Lin, C.-T.; Tseng, K.-C.; Wu, J.-S.; Juang, R.-H. Site-specific phosphorylation of L-form starch phosphorylase by the protein kinase activity from sweet potato roots. Planta 2005, 223, 468–478.
- Mori, H.; Tanizawa, K.; Fukui, T. A chimeric alpha-glucan phosphorylase of plant type L and H isozymes. Functional role of 78-residue insertion in type L isozyme. J. Biol. Chem. 1993, 268, 5574–5581.
- Liddle, A.M.; Manners, D.J.; Wright, A. Studies on carbohydrate-metabolizing enzymes. 6. The action of potato phosphorylase (P-enzyme) on starch-type polysaccharides. Biochem. J. 1961, 80, 304–309.
- Shimomura, S.; Nagai, M.; Fukui, T. Comparative Glucan Specificities of Two Types of Spinach Leaf Phosphorylase. J. Biochem. 1982, 91, 703–717.
- Young, N.D.; Debellé, F.; Oldroyd, G.E.D.; Geurts, R.; Cannon, S.B.; Udvardi, M.; Benedito, V.A.; Mayer, K.; Gouzy, J.; Schoof, H.; et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nat. Cell Biol. 2011, 480, 520–524.
- The Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641.
- Chandler, V.L.; Brendel, V. The Maize Genome Sequencing Project. Plant Physiol. 2002, 130, 1594–1597.
- International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 2010, 463, 763–768.
- Nakano, K.; Mori, H.; Fukui, T. Molecular Cloning of cDNA Encoding Potato Amyloplast a-Glucan Phosphorylase and the Structure of Its Transit Peptide. J. Biochem. 1989, 106, 691–695.
- Camirand, A.; St-Pierre, B.; Marineau, C.; Brisson, N. Occurrence of a copia-like transposable element in one of the introns of the potato starch phosphorylase gene. Mol. Genet. Genom. 1990, 224, 33–39.
- Lin, C.-T.; Yeh, K.-W.; Lee, P.-D.; Su, J.-C. Primary Structure of Sweet Potato Starch Phosphorylase Deduced from its cDNA Sequence. Plant Physiol. 1991, 95, 1250–1253.
- Kang, Y.J.; Kim, S.K.; Kim, M.Y.; Lestari, P.; Kim, K.H.; Ha, B.-K.; Jun, T.H.; Hwang, W.J.; Lee, T.; Lee, J.; et al. Genome sequence of mungbean and insights into evolution within Vigna species. Nat. Commun. 2014, 5, 5443.
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964.
- Slugina, M.A.; Shchennikova, A.V.; Kochieva, E. The expression pattern of the Pho1a genes encoding plastidic starch phosphorylase correlates with the degradation of starch during fruit ripening in green-fruited and red-fruited tomato species. Funct. Plant Biol. 2019, 46, 1146.
- Walley, J.W.; Shen, Z.; Sartor, R.; Wu, K.J.; Osborn, J.; Smith, L.G.; Briggs, S.P. Reconstruction of protein networks from an atlas of maize seed proteotypes. Proc. Natl. Acad. Sci. USA 2013, 110, E4808–E4817.
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203.




