
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Natsuko Chiba | + 2138 word(s) | 2138 | 2020-07-27 11:02:25 | | | |
| 2 | Rita Xu | -675 word(s) | 1463 | 2020-08-03 05:15:24 | | |
Video Upload Options
Breast cancer gene 1 (BRCA1) is a tumor suppressor associated with hereditary breast and ovarian cancer and forms a heterodimer with BRCA1-associated RING domain protein 1 (BARD1). Centrosomes are the major microtubule-organizing centers in animal cells and are critical for the formation of a bipolar mitotic spindle. BRCA1 and BARD1 localize to the centrosome during the cell cycle, and the BRCA1/BARD1 dimer ubiquitinates centrosomal proteins to regulate centrosome function.
1. Definition
Breast cancer gene 1 (BRCA1) is a tumor suppressor associated with hereditary breast and ovarian cancer and forms a heterodimer with BRCA1-associated RING domain protein 1 (BARD1). BRCA1/BARD1 functions in multiple cellular processes including DNA repair and centrosome regulation. Centrosomes are the major microtubule-organizing centers in animal cells and are critical for the formation of a bipolar mitotic spindle. BRCA1 and BARD1 localize to the centrosome during the cell cycle, and the BRCA1/BARD1 dimer ubiquitinates centrosomal proteins to regulate centrosome function.
2. Introduction
Germline mutations in Breast Cancer gene 1 (BRCA1) are associated with familial breast and ovarian cancers [1]. In women with BRCA1 mutations, the risk of developing breast cancer by the age of 70 years is 57% and that of ovarian cancer is 40% [2]. BRCA1 has a RING domain in the amino (N)-terminal region and two BRCT domains in the carboxy (C)-terminal region (Figure 1). BRCA1-associated RING domain 1 (BARD1) was identified as a binding protein of the N-terminal region (amino acid (aa) 1–304) of BRCA1 by yeast two-hybrid screening [3]. BARD1 contains an N-terminal RING domain, three tandem ankyrin (ANK) repeats, and two BRCT domains (Figure 1). BARD1 forms a heterodimer with BRCA1 via their RING domain, and the C-terminal region of BRCA1 contributes markedly to the stability of the heterodimer [4][5]. The BRCA1/BARD1 dimer is involved in DNA repair, centrosome regulation, chromatin remodeling, and transcription [6].
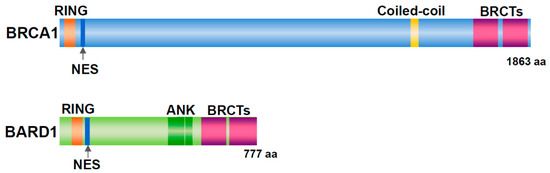
Figure 1. Structure of BRCA1 and BARD1. Both proteins have a RING domain and nuclear export signal (NES) in the N-terminal region and two BRCT domains in the C-terminal region. In addition, BRCA1 includes a coiled-coil domain. BARD1 contains three ankyrin (ANK) repeats.
Centrosomes are the major microtubule (MT)-organizing centers (MTOC) in animal cells; they control cell shape, polarity, and motility, and mediate the formation of a bipolar mitotic spindle [7][8]. Each centrosome consists of a pair of centrioles called the mother and daughter centrioles, surrounded by a protein matrix known as the pericentriolar matrix (PCM) (Figure 2A). The PCM contains γ-tubulin ring complexes (γ-TuRCs) that play important roles in nucleating, anchoring, and positioning MTs. The single centrosome in the G1 phase duplicates only once per cell cycle (in S phase), and one centrosome is inherited by each daughter cell [8]. Centrosome duplication is precisely controlled by centriole duplication during the cell cycle (Figure 2B). Centrosome duplication is initiated by the physical separation of a pair of centrioles (centriole disengagement) in late mitosis-early G1 phase. The new daughter centriole starts to form a procentriole perpendicular to each mother centriole in early S phase. Each daughter centriole gradually elongates during the S and G2 phases. In late G2 phase, the two centrosomes separate and migrate to form the two opposing poles of the mitotic spindle [9].
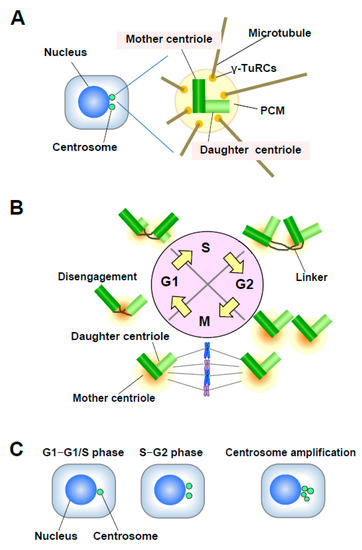
Figure 2. (A) Structure of the centrosome. The centrosome consists of a pair of centrioles, mother centriole and daughter centriole, embedded in the pericentriolar matrix (PCM). The PCM contains γ-TuRCs, which play roles in nucleating, anchoring, and positioning microtubules (MTs). (B) Centrosome duplication in the cell cycle. The mother and daughter centrioles are disengaged in late mitosis-early G1 phase. After centriole disengagement, a proteinaceous linker is established between the two centrioles and physically connects them. The building of the new centriole starts in the early S phase with the formation of a procentriole at each centriole. One new daughter centriole forms perpendicularly to each mother centriole during the S phase, and the new daughter centriole gradually elongates during the S and G2 phases. In late G2 phase, the two centrosomes separate through the dissolution of the linker and move to opposite sides of the cell to form the spindle poles. (C) Number of centrosomes. Normally, the centrosome number is one or two in interphase. Centrosome amplification is usually defined as more than two centrosomes per cell.
BARD1 localizes to the centrosome and functions in centrosome duplication and the regulation of MT organizing activity of centrosomes together with BRCA1 [10][11][12]. We recently identified the BRCA1/BARD1-interacting proteins Obg-like ATPase 1 (OLA1) and receptor for activated C kinase (RACK1) [13][14].
3. The BRCA1/BARD1 Heterodimer Functions in Centrosome Regulation
BRCA1 and BARD1 localize to the centrosome throughout the cell cycle [11][12][15]. Two regions of BRCA1, aa 504–803 and aa 802–1002, mediate its binding to γ-tubulin [16][17]. Brodie et al. reported that both N and C-terminal regions of BRCA1, but not the RING domain, are required for its centrosomal localization, independently of BARD1 and γ-tubulin [18]. Tarapore et al. reported that only the middle portion of BRCA1, namely aa 802–1002, is responsible for its localization to centrosomes [17]. The N-terminal nuclear export sequence (NES) of BRCA1 is important for targeting, turnover, and function at the centrosome, suggesting regulation by chromosome region maintenance 1 (CRM1). In addition, the mitotic kinase Aurora A contributes to BRCA1 retention at the centrosome [18]. Inside the centrosome, BRCA1 localizes to mother centrioles, whereas daughter centrioles acquire BRCA1 prior to the initiation of procentriole formation in late G1 phase [17].
Similar to BRCA1, the N- and C-terminal regions of BARD1, but not the RING domain, are critical for its centrosomal localization independently of BRCA1 [19]. The N-terminal NES mediates the centrosomal localization of BARD1, suggesting that the CRM1 is also involved in this process. The RING domains are not necessary for the centrosomal localization of BRCA1 and BARD1. Fluorescence recovery after photobleaching assays indicate that the retained centrosomal pool of BARD1 is half the amount observed for BRCA1, and that BARD1 is one of the most highly mobile proteins in the centrosome [19].
The RING domains of BRCA1 and BARD1 have E3 ubiquitin ligase activity, which increases dramatically in the BRCA1/BARD1 RING domain heterodimer [20]. Several cancer-associated BRCA1 RING domain variants abolish binding to BARD1 and the E3 ubiquitin ligase activity [21][22]. The BRCA1/BARD1 dimer ubiquitinates centrosomal proteins, including γ-tubulin, nucleophosmin/B23 (NPM1), and receptor for hyaluronan (HA)-mediated motility (RHAMM)/hyaluronan-mediated motility receptor (HMMR) [10][23][24].
BRCA1/BARD1 monoubiquitinates γ-tubulin at lysines K48 and K344, and the C-terminal region of BRCA1 is required for this function [10]. Suppression or overexpression of BRCA1 or BARD1 results in centrosome amplification in mammary tissue-derived cells [10][25]. Centrosome amplification induced by BRCA1 suppression is caused by premature centriole disengagement and centriole reduplication [26]. These findings suggest that BRCA1 functions in the regulation of centrosome number by controlling centriole duplication in mammary cells. Furthermore, BRCA1/BARD1 inhibits centrosome-dependent MT organizing activity, and the C-terminal region of BARD1 is necessary for the inhibition [11][12]. MT organizing activity was analyzed by detecting aster formation by centrosomes the in vivo MT regrowth assay and in vitro assay. BRCA1/BARD1 E3 ubiquitin ligase activity and its inhibitory effect on MT aster formation are suppressed by Aurora A and promoted by protein phosphatase 1α [27]. Aurora A is a mitotic kinase, and its overexpression causes centrosome amplification in cells [28]. The E3 ubiquitin ligase activity of BRCA1/BARD1 is important for both functions, regulation of centriole duplication and the inhibitory effect on MT aster formation. Monoubiquitination of γ-tubulin at K344 is critical for both functions, whereas that at K48 functions only in centrosome duplication [11]. Partially consistent with these findings, embryonic fibroblasts, which are not mammary cells, from BRCA1-knockout mice, show centrosome amplification [29]. In MT organization by the centrosome, MT nucleation is initiated by the γTuRC, and then the MT anchoring complex at the sub-distal appendages in mother centrioles anchors the MT-nucleated γTuRC. The nucleated MTs then elongate to form MT asters. Terapore et al. analyzed MT nucleation and MT anchoring and/or elongation at the centrosome separately, and concluded that BRCA1 suppresses MT anchoring and/or elongation but not MT nucleation [17].
NPM1 interacts with the N-terminal region of BRCA1 and BARD1 in a manner dependent on BRCA1/BARD1 heterodimer formation and is polyubiquitinated by BRCA1/BARD1, resulting in its stabilization. In mitotic cells, NPM1 colocalizes with BARD1 at chromosomal surfaces and the perichromosomal cytoplasm, and with BRCA1 at the spindle poles [23].
RHAMM, a member of the transforming acidic coiled-coil (TACC) family, localizes to the centrosome and associates with MTs [30][31]. Similar to the effects of BRCA1 inhibition, depletion of RHAMM causes centrosome amplification in mammary tissue-derived cells. RHAMM is associated with BRCA1, BRCA2, and Aurora A, and is polyubiquitinated and stabilized by BRCA1/BARD1 [24]. The RHAMM ortholog in Xenopus laevis, XRHAMM, regulates spindle pole assembly mediated by the BRCA1/BARD1 heterodimer [32]. A yeast two-hybrid screening identified the association between the TAC-1 and BRD-1 C. elegans proteins, which are orthologs of TACCs and BARD1, respectively [33][34].
References
- Miki, Y.; Swensen, J.; Shattuck-Eidens, D.; Futreal, P.A.; Harshman, K.; Tavtigian, S.; Liu, Q.; Cochran, C.; Bennett, L.M.; Ding, W. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994, 266, 66–71.
- Chen, S.; Parmigiani, G. Meta-analysis of BRCA1 and BRCA2 penetrance. J. Clin. Oncol. 2007, 25, 1329–1333.
- Wu, L.C.; Wang, Z.W.; Tsan, J.T.; Spillman, M.A.; Phung, A.; Xu, X.L.; Yang, M.C.W.; Hwang, L.Y.; Bowcock, A.M.; Baer, R. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat. Genet. 1996, 14, 430–440.
- Brzovic, P.S.; Rajagopal, P.; Hoyt, D.W.; King, M.C.; Klevit, R.E. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol. 2001, 8, 833–837.
- Simons, A.M.; Horwitz, A.A.; Starita, L.M.; Griffin, K.; Williams, R.S.; Glover, J.M.; Parvin, J.D. BRCA1 DNA-binding activity is stimulated by BARD1. Cancer Res. 2006, 66, 2012–2018.
- Takaoka, M.; Miki, Y. BRCA1 gene: Function and deficiency. Int. J. Clin. Oncol. 2018, 23, 36–44.
- Conduit, P.T.; Wainman, A.; Raff, J.W. Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell Biol. 2015, 16, 611–624.
- Nigg, E.A.; Holland, A.J. Once and only once: Mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 297–312.
- Fujita, H.; Yoshino, Y.; Chiba, N. Regulation of the centrosome cycle. Mol. Cell. Oncol. 2016, 3, e1075643.
- Starita, L.M.; Machida, Y.; Sankaran, S.; Elias, J.E.; Griffin, K.; Schlegel, B.P.; Gygi, S.P.; Parvin, J.D. BRCA1-dependent ubiquitination of gamma-tubulin regulates centrosome number. Mol. Cell. Oncol. 2004, 24, 8457–8466.
- Sankaran, S.; Starita, L.M.; Groen, A.C.; Ko, M.J.; Parvin, J.D. Centrosomal microtubule nucleation activity is inhibited by BRCA1-dependent ubiquitination. Mol. Cell. Oncol. 2005, 25, 8656–8668.
- Sankaran, S.; Starita, L.M.; Simons, A.M.; Parvin, J.D. Identification of domains of BRCA1 critical for the ubiquitin-dependent inhibition of centrosome function. Cancer Res. 2006, 66, 4100–4107.
- Matsuzawa, A.; Kanno, S.; Nakayama, M.; Mochiduki, H.; Wei, L.; Shimaoka, T.; Furukawa, Y.; Kato, K.; Shibata, S.; Yasui, A.; et al. The BRCA1/BARD1-Interacting Protein OLA1 Functions in Centrosome Regulation. Mol. Cell 2014, 53, 101–114.
- Yoshino, Y.; Qi, H.; Kanazawa, R.; Sugamata, M.; Suzuki, K.; Kobayashi, A.; Shindo, K.; Matsuzawa, A.; Shibata, S.; Endo, S.; et al. RACK1 regulates centriole duplication by controlling localization of BRCA1 to the centrosome in mammary tissue-derived cells. Oncogene 2019, 38, 3077–3092.
- Hsu, L.C.; White, R.L. BRCA1 is associated with the centrosome during mitosis. Proc. Natl. Acad. Sci. USA 1998, 95, 12983–12988.
- Hsu, L.C.; Doan, T.P.; White, R.L. Identification of a gamma-tubulin-binding domain in BRCA1. Cancer Res. 2001, 61, 7713–7718.
- Tarapore, P.; Hanashiro, K.; Fukasawa, K. Analysis of centrosome localization of BRCA1 and its activity in suppressing centrosomal aster formation. Cell Cycle 2012, 11, 2931–2946.
- Brodie, K.M.; Henderson, B.R. Characterization of BRCA1 protein targeting, dynamics, and function at the centrosome: A role for the nuclear export signal, CRM1, and Aurora A kinase. J. Biol. Chem. 2012, 287, 7701–7716.
- Brodie, K.M.; Mok, M.T.; Henderson, B.R. Characterization of BARD1 targeting and dynamics at the centrosome: The role of CRM1, BRCA1 and the Q564H mutation. Cell. Signal. 2012, 24, 451–459.
- Hashizume, R.; Fukuda, M.; Maeda, I.; Nishikawa, H.; Oyake, D.; Yabuki, Y.; Ogata, H.; Ohta, T. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem. 2001, 276, 14537–14540.
- Brzovic, P.S.; Keeffe, J.R.; Nishikawa, H.; Miyamoto, K.; Fox, D.; Fukuda, M., III; Ohta, T.; Klevit, R. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc. Natl. Acad. Sci. USA 2003, 100, 5646–5651.
- Ransburgh, D.J.; Chiba, N.; Ishioka, C.; Toland, A.E.; Parvin, J.D. Identification of breast tumor mutations in BRCA1 that abolish its function in homologous DNA recombination. Cancer Res. 2010, 70, 988–995.
- Sato, K.; Hayami, R.; Wu, W.; Nishikawa, T.; Nishikawa, H.; Okuda, Y.; Ogata, H.; Fukuda, M.; Ohta, T. Nucleophosmin/B23 is a candidate substrate for the BRCA1-BARD1 ubiquitin ligase. J. Biol. Chem. 2004, 279, 30919–30922.
- Pujana, M.A.; Han, J.D.; Starita, L.M.; Stevens, K.N.; Tewari, M.; Ahn, J.S.; Rennert, G.; Moreno, V.; Kirchhoff, T.; Gold, B.; et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat. Genet. 2007, 39, 1338–1349.
- Yoshino, Y.; Qi, H.; Fujita, H.; Shirota, M.; Abe, S.; Komiyama, Y.; Shindo, K.; Nakayama, M.; Matsuzawa, A.; Kobayashi, A.; et al. BRCA1-interacting Protein OLA1 Requires Interaction with BARD1 to Regulate Centrosome Number. Mol. Cancer Res. 2018, 16, 1499–1511.
- Ko, M.J.; Murata, K.; Hwang, D.S.; Parvin, J.D. Inhibition of BRCA1 in breast cell lines causes the centrosome duplication cycle to be disconnected from the cell cycle. Oncogene 2006, 25, 298–303.
- Sankaran, S.; Crone, D.E.; Palazzo, R.E.; Parvin, J.D. BRCA1 regulates gamma-tubulin binding to centrosomes. Cancer Biol. Ther. 2007, 6, 1853–1857.
- Lukasiewicz, K.B.; Lingle, W.L. Aurora A, centrosome structure, and the centrosome cycle. Environ. Mol. Mutagen. 2009, 50, 602–619.
- Xu, X.; Weaver, Z.; Linke, S.P.; Li, C.; Gotay, J.; Wang, X.W.; Harris, C.C.; Ried, T.; Deng, C.X. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell 1999, 3, 389–395.
- Peset, I.; Vernos, I. The TACC proteins: TACC-ling microtubule dynamics and centrosome function. Trends Cell Biol. 2008, 18, 379–388.
- Maxwell, C.A.; Keats, J.J.; Crainie, M.; Sun, X.; Yen, T.; Shibuya, E.; Hendzel, M.; Chan, G.; Pilarski, L.M. RHAMM is a centrosomal protein that interacts with dynein and maintains spindle pole stability. Mol. Biol. Cell 2003, 14, 2262–2276.
- Joukov, V.; Groen, A.C.; Prokhorova, T.; Gerson, R.; White, E.; Rodriguez, A.; Walter, J.C.; Livingston, D.M. The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell 2006, 127, 539–552.
- Li, S.; Armstrong, C.M.; Bertin, N.; Ge, H.; Milstein, S.; Boxem, M.; Vidalain, P.O.; Han, J.D.J.; Chesneau, A.; Hao, T.; et al. A map of the interactome network of the metazoan C. elegans. Science 2004, 303, 540–543.
- Boulton, S.J.; Martin, J.S.; Polanowska, J.; Hill, D.E.; Gartner, A.; Vidal, M. BRCA1/BARD1 orthologs required for DNA repair in Caenorhabditis elegans. Curr. Biol. 2004, 14, 33–39.




