
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Zhang Hua | + 2330 word(s) | 2330 | 2021-09-28 08:50:29 |
Video Upload Options
Rolling circle amplification (RCA) is an isothermal enzymatic process in which a short nucleic acid primer is amplified to form a long single-stranded nucleic acid using a circular template and special nucleic acid polymerases. Furthermore, this approach can be further engineered into a device for point-of-need monitoring of environmental pollutants.
1. Introduction
In recent years, the discharge of contaminants from industrial and agricultural activities and urban wastewater has caused serious contamination of the aqueous system, posing a great potential threat to human health and aquatic life. These contaminants can be divided into three categories: (i) inorganic chemical substances, (ii) organic pollutants and (iii) microorganisms. These substances can cause adverse effects on the environment [1][2][3][4], for example, the disruption of hormones and the endocrine system and the induction of cytotoxicity and/or genotoxicity and carcinogenesis [5][6]. The variable composition of pollutants and their location in aqueous environments over time have resulted in increasing focus on new technologies that use cheap and real-time strategies to monitor pollutants. Most of these strategies are based on laboratory platforms, such as inductively coupled plasma mass spectrometry (ICP-MS) for the detection of heavy metal ions, liquid chromatography-tandem mass spectrometry (LC-MS) for the detection of small organic chemicals or their metabolites, and polymerase chain reaction (PCR) for the detection of nucleic acids and genetic information, which require preprocessing and frequent data sampling, which means that they are both expensive and slow. These aspects highlight the need to develop a new strategy that is more sensitive, portable, and efficient for on-site detection of pollutants composed of multiple substances [7][8][9][10].
Recently, rolling circle amplification (RCA)-based analytical methods have received increasing attention in environmental monitoring. RCA is an uncomplicated and efficient isothermal enzymatic process using unique DNA and RNA polymerases to produce long single-stranded DNA (ssDNA) and RNA [11][12]. In RCA, the polymerase will spontaneously and continuously add nucleotides to the primers that bind to the circular template, generating long ssDNA with tandem repeats of tens to hundreds of orders of magnitude. Unlike PCR, which requires a thermal cycler and thermostable DNA polymerase. RCA can be in solution, on a solid support, or in a complex biological environment at a constant temperature (room temperature to 37 °C). The ability of RCA to grow a long DNA chain on a solid support or inside a cell from one molecular binding event enables the detection of targets at a single molecule level [13][14][15]. In addition, an RCA product comprising repeating cyclic sequences complementary to template DNA can be customized by template design. By designing the template, the customizable DNA product includes functional sequences, including aptamers, DNAzymes, spacer domains, and restriction endonuclease sites. Of course, by hybridizing the RCA product with a complementary nucleic acid linked to a functional part including biotin [16][17], fluorophores [18][19], antibodies [20], and nanoparticles [21][22][23][24], it is easy to synthesize a multifunctional material with a variety of properties, including biorecognition and biosensing. Collectively, the properties of high-efficiency isothermal amplification, single-molecule sensitivity, versatility of structure and composition, and multivalences make RCA a powerful tool in aqueous environments [25][26][27]. Currently, RCA has been extensively studied to develop sensitive methods for detecting DNA, RNA, DNA methylation, single nucleotide polymorphisms, small molecules, proteins, and cells. In addition to diagnosis, RCA has also been proven to be effective for cell-free cloning and sequencing [28][29], in situ genotyping and genome-wide analysis of cells and tissues [30][31][32][33][34]. Recently, RCA has received widespread attention for its use in the production of DNA nanostructures such as origami, nanoribbons, nanotubes, DNA nanoscaffolds, and DNA metamaterials for periodic nanocomponents [11][35][36][37][38]. Importantly, these materials have high prospects in a wide range of applications, including environmental monitoring, drug delivery, and in vivo imaging of manufacturing electronic circuits, including DNA-based materials.
2. Advantages and Disadvantages of the RCA Assay
2.1. Fundamentals of RCA
| Features | Conventional PCR Assay | Real Time-PCR Assay | RCA Assay |
|---|---|---|---|
| Sensitivity | Sensitive | Highly sensitive | Highly sensitive |
| Specificity | Specific | Specific | Specific |
| Temperature conditions | Thermal cycle | Thermal cycle | Isothermal |
| Inhibition by biological samples | Yes | Yes | No |
| Instruments required | Thermocycler | Thermocycler | Not required |
| Post-assay analysis | Required | Required | Generally not required |
| Amplicon detection methods | Gel electrophoresis | Real-time detection/amplification graph | Gel electrophoresis, Turbidity measurement by visual inspection or using a real-time turbidimeter; dye-based visual detection |
| Qualitative detection | Yes | Yes | Yes |
| Quantitative detection | No | Yes | Semi-quantitative |
| Portability | Partially | Yes | Yes |
| Overall assay time | 3–5 h | 2.5–4 h | 1–1.5 h |
| Cost effectiveness | Less expensive | Expensive | Less expensive |
2.2. Exponential RCA Amplification
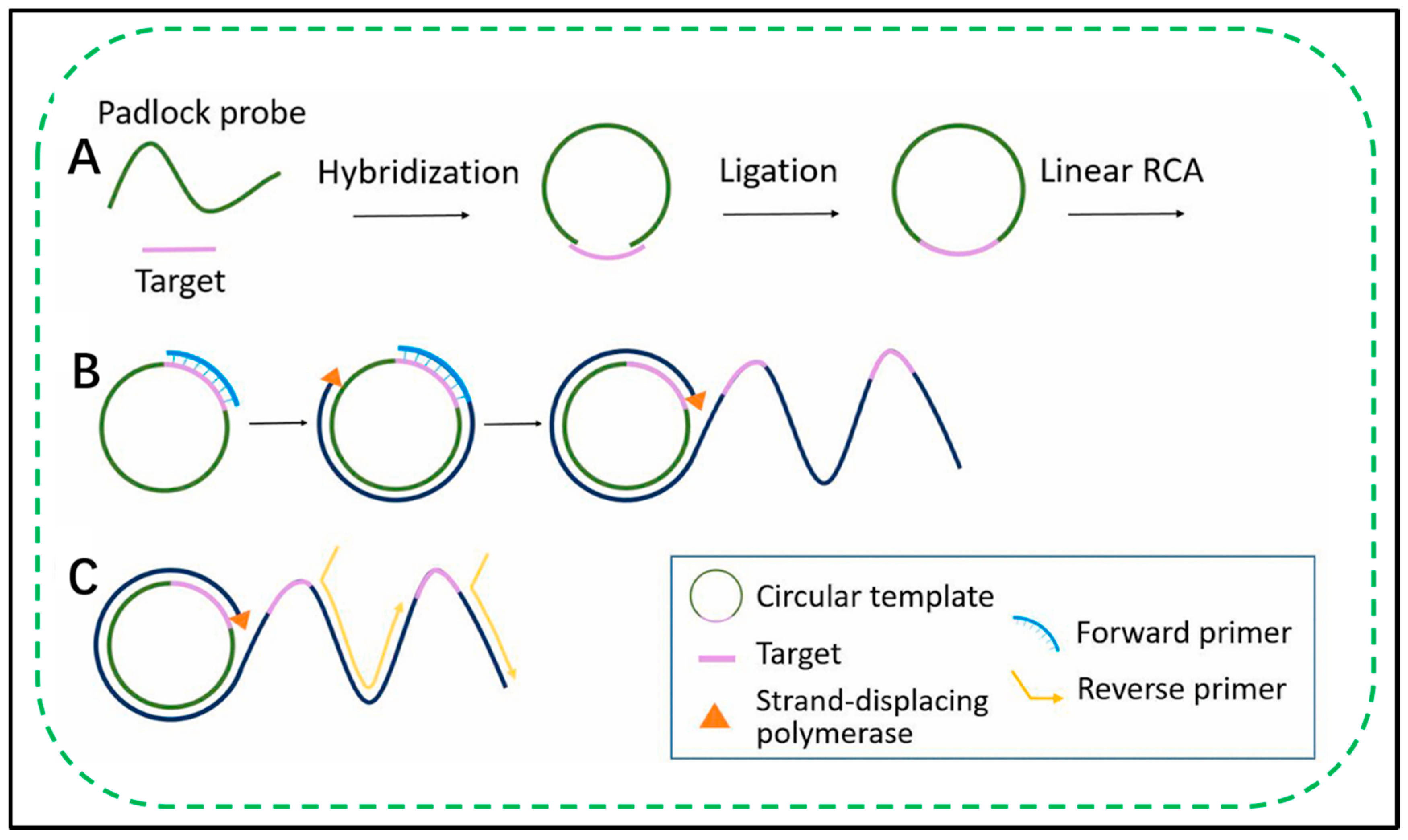
2.3. Detection of the RCA Product
3. Conclusions and Perspective
References
- Blair, B.D.; Crago, J.P.; Hedman, C.J.; Klaper, R.D. Pharmaceuticals and personal care products found in the Great Lakes above concentrations of environmental concern. Chemosphere 2013, 93, 2116–2123.
- Eckert, E.M.; Di Cesare, A.; Kettner, M.T.; Arias-Andres, M.; Fontaneto, D.; Grossart, H.P.; Corno, G. Microplastics increase impact of treated wastewater on freshwater microbial community. Environ. Pollut. 2018, 234, 495–502.
- Wang, Z.; Han, S.; Cai, M.; Du, P.; Zhang, Z.; Li, X. Environmental behaviour of methamphetamine and ketamine in aquatic ecosystem: Degradation, bioaccumulation, distribution, and associated shift in toxicity and bacterial community. Water Res. 2020, 174, 115585.
- Yu, Y.Y.; Huang, Q.X.; Wang, Z.F.; Zhang, K.; Tang, C.M.; Cui, J.L.; Feng, J.L.; Peng, X.Z. Occurrence and behaviour of pharmaceuticals, steroid hormones, and endocrine-disrupting personal care products in wastewater and the recipient river water of the Pearl River Delta, South China. J. Environ. Monitor. 2011, 13, 871–878.
- Cheng, D.L.; Ngo, H.H.; Guo, W.S.; Liu, Y.W.; Zhou, J.L.; Chang, S.W.; Nguyen, D.D.; Bui, X.T.; Zhang, X.B. Bioprocessing for elimination antibiotics and hormones from swine wastewater. Sci. Total Environ. 2018, 621, 1664–1682.
- Hamid, H.; Eskicioglu, C. Fate of oestrogenic hormones in wastewater and sludge treatment: A review of properties and analytical detection techniques in sludge matrix. Water Res. 2012, 46, 5813–5833.
- Ganjali, M.R.; Faridbod, F.; Davarkhah, N.; Shahtaheri, S.J.; Norouzi, P. All Solid State Graphene Based Potentiometric Sensors for Monitoring of Mercury Ions in Waste Water Samples. Int. J. Environ. Res. 2015, 9, 333–340.
- Gavrilescu, M.; Demnerova, K.; Aamand, J.; Agathoss, S.; Fava, F. Emerging pollutants in the environment: Present and future challenges in biomonitoring, ecological risks and bioremediation. New Biotechnol. 2015, 32, 147–156.
- Henderson, R.K.; Baker, A.; Murphy, K.R.; Hamblya, A.; Stuetz, R.M.; Khan, S.J. Fluorescence as a potential monitoring tool for recycled water systems: A review. Water Res. 2009, 43, 863–881.
- Sousa, J.C.G.; Ribeiro, A.R.; Barbosa, M.O.; Pereira, M.F.R.; Silva, A.M.T. A review on environmental monitoring of water organic pollutants identified by EU guidelines. J. Hazard. Mater. 2018, 344, 146–162.
- Ali, M.M.; Li, F.; Zhang, Z.; Zhang, K.; Kang, D.-K.; Ankrum, J.A.; Le, X.C.; Zhao, W. Rolling circle amplification: A versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014, 43, 3324–3341.
- Lizardi, P.M.; Huang, X.H.; Zhu, Z.R.; Bray-Ward, P.; Thomas, D.C.; Ward, D.C. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 1998, 19, 225–232.
- Murakami, T.; Sumaoka, J.; Komiyama, M. Sensitive isothermal detection of nucleic-acid sequence by primer generation-rolling circle amplification. Nucleic Acids Res. 2009, 37, e19.
- Zhao, W.A.; Ali, M.M.; Brook, M.A.; Li, Y.F. Rolling circle amplification: Applications in nanotechnology and biodetection with functional nucleic acids. Angew. Chem. Int. Ed. 2008, 47, 6330–6337.
- Zhao, Y.X.; Chen, F.; Li, Q.; Wang, L.H.; Fan, C.H. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545.
- Guo, Y.N.; Wang, Y.; Liu, S.; Yu, J.H.; Wang, H.Z.; Wang, Y.L.; Huang, J.D. Label-free and highly sensitive electrochemical detection of E-coli based on rolling circle amplifications coupled peroxidase-mimicking DNAzyme amplification. Biosens. Bioelectron. 2016, 75, 315–319.
- Zhang, K.Y.; Lv, S.Z.; Lu, M.H.; Tang, D.P. Photoelectrochemical biosensing of disease marker on p-type Cu-doped Zn0.3Cd0.7S based on RCA and exonuclease III amplification. Biosens. Bioelectron. 2018, 117, 590–596.
- Qiu, Z.L.; Shu, J.; He, Y.; Lin, Z.Z.; Zhang, K.Y.; Lv, S.Z.; Tang, D.P. CdTe/CdSe quantum dot-based fluorescent aptasensor with hemin/G-quadruplex DNzyme for sensitive detection of lysozyme using rolling circle amplification and strand hybridization. Biosens. Bioelectron. 2017, 87, 18–24.
- Sun, D.P.; Lu, J.; Luo, Z.F.; Zhang, L.Y.; Liu, P.Q.; Chen, Z.G. Competitive electrochemical platform for ultrasensitive cytosensing of liver cancer cells by using nanotetrahedra structure with rolling circle amplification. Biosens. Bioelectron. 2018, 120, 8–14.
- Zhang, K.Y.; Lv, S.Z.; Lin, Z.Z.; Li, M.J.; Tang, D.P. Bio-bar-code-based photoelectrochemical immunoassay for sensitive detection of prostate-specific antigen using rolling circle amplification and enzymatic biocatalytic precipitation. Biosens. Bioelectron. 2018, 101, 159–166.
- Chen, A.Y.; Ma, S.Y.; Zhuo, Y.; Chai, Y.Q.; Yuan, R. In Situ Electrochemical Generation of Electrochemiluminescent Silver Naonoclusters on Target-Cycling Synchronized Rolling Circle Amplification Platform for MicroRNA Detection. Anal. Chem. 2016, 88, 3203–3210.
- Fan, T.; Du, Y.; Yao, Y.; Wu, J.; Meng, S.; Luo, J.; Zhang, X.; Yang, D.; Wang, C.; Qian, Y.; et al. Rolling circle amplification triggered poly adenine-gold nanoparticles production for label-free electrochemical detection of thrombin. Sens. Actuators B Chem. 2018, 266, 9–18.
- He, Y.; Yang, X.; Yuan, R.; Chai, Y.Q. “Off” to “On” Surface-Enhanced Raman Spectroscopy Platform with Padlock Probe-Based Exponential Rolling Circle Amplification for Ultrasensitive Detection of MicroRNA 155. Anal. Chem. 2017, 89, 2866–2872.
- Qiu, Z.L.; Shu, J.; Tang, D.P. Near-Infrared-to-Ultraviolet Light-Mediated Photoelectrochemical Aptasensing Platform for Cancer Biomarker Based on Core Shell NaYF4:Yb, Upconversion Microrods. Anal. Chem. 2018, 90, 1021–1028.
- Kim, T.Y.; Lim, M.C.; Woo, M.A.; Jun, B.H. Radial Flow Assay Using Gold Nanoparticles and Rolling Circle Amplification to Detect Mercuric Ions. Nanomaterials 2018, 8, 81.
- Wen, J.; Li, W.S.; Li, J.Q.; Tao, B.B.; Xu, Y.Q.; Li, H.J.; Lu, A.P.; Sun, S.G. Study on rolling circle amplification of Ebola virus and fluorescence detection based on graphene oxide. Sens. Actuators B Chem. 2016, 227, 655–659.
- Zhao, J.; Lei, Y.-M.; Chai, Y.-Q.; Yuan, R.; Zhuo, Y. Novel electrochemiluminescence of perylene derivative and its application to mercury ion detection based on a dual amplification strategy. Biosens. Bioelectron. 2016, 86, 720–727.
- Osborne, R.J.; Thornton, C.A. Cell-free cloning of highly expanded CTG repeats by amplification of dimerized expanded repeats. Nucleic Acids Res. 2008, 36, e24.
- Wang, F.; Lu, C.H.; Liu, X.Q.; Freage, L.; Willner, I. Amplified and Multiplexed Detection of DNA Using the Dendritic Rolling Circle Amplified Synthesis of DNAzyme Reporter Units. Anal. Chem. 2014, 86, 1614–1621.
- Du, Y.C.; Zhu, Y.J.; Li, X.Y.; Kong, D.M. Amplified detection of genome-containing biological targets using terminal deoxynucleotidyl transferase-assisted rolling circle amplification. Chem. Commun. 2018, 54, 682–685.
- Inoue, J.; Shigemori, Y.; Mikawa, T. Improvements of rolling circle amplification (RCA) efficiency and accuracy using Thermus thermophilus SSB mutant protein. Nucleic Acids Res. 2006, 34, e69.
- Li, J.S.; Deng, T.; Chu, X.; Yang, R.H.; Jiang, J.H.; Shen, G.L.; Yu, R.Q. Rolling Circle Amplification Combined with Gold Nanoparticle Aggregates for Highly Sensitive Identification of Single-Nucleotide Polymorphisms. Anal. Chem. 2010, 82, 2811–2816.
- Qi, X.Q.; Bakht, S.; Devos, K.M.; Gale, M.D.; Osbourn, A. L-RCA (ligation-rolling circle amplification): A general method for genotyping of shingle nucleotide polymorphisms (SNPs). Nucleic Acids Res. 2001, 29, e116.
- Zhou, H.X.; Wang, H.; Liu, C.H.; Wang, H.H.; Duan, X.R.; Li, Z.P. Ultrasensitive genotyping with target-specifically generated circular DNA templates and RNA FRET probes. Chem. Commun. 2015, 51, 11556–11559.
- Ko, O.; Han, S.; Lee, J.B. Selective release of DNA nanostructures from DNA hydrogel. J. Ind. Eng. Chem. 2020, 84, 46–51.
- Liu, M.; Zhang, Q.; Li, Z.P.; Gu, J.; Brennan, J.D.; Li, Y.F. Programming a topologically constrained DNA nanostructure into a sensor. Nat. Commun. 2016, 7, 12074.
- Xu, H.; Zhang, S.X.; Ouyang, C.H.; Wang, Z.M.; Wu, D.; Liu, Y.Y.; Jiang, Y.F.; Wu, Z.S. DNA nanostructures from palindromic rolling circle amplification for the fluorescent detection of cancer-related microRNAs. Talanta 2019, 192, 175–181.
- Zhang, Z.Q.; Zhang, H.Z.; Wang, F.; Zhang, G.D.; Zhou, T.; Wang, X.F.; Liu, S.Z.; Liu, T.T. DNA Block Macromolecules Based on Rolling Circle Amplification Act as Scaffolds to Build Large-Scale Origami Nanostructures. Macromol. Rapid. Commun. 2018, 39, 1800263.
- Mohsen, M.G.; Kool, E.T. The Discovery of Rolling Circle Amplification and Rolling Circle Transcription. Acc. Chem. Res. 2016, 49, 2540–2550.
- Fire, A.; Xu, S.Q. Rolling Replication of Short Dna Circles. Proc. Natl. Acad. Sci. USA 1995, 92, 4641–4645.
- Liu, D.Y.; Daubendiek, S.L.; Zillman, M.A.; Ryan, K.; Kool, E.T. Rolling circle DNA synthesis: Small circular oligonucleotides as efficient templates for DNA polymerases. J. Am. Chem. Soc. 1996, 118, 1587–1594.
- Blanco, L.; Bernad, A.; Lazaro, J.M.; Martin, G.; Garmendia, C.; Salas, M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J. Biol. Chem. 1989, 264, 8935–8940.
- Krzywkowski, T.; Kuhnemund, M.; Wu, D.; Nilsson, M. Limited reverse transcriptase activity of phi29 DNA polymerase. Nucleic Acids Res. 2018, 46, 3625–3632.
- Neumann, F.; Hernandez-Neuta, I.; Grabbe, M.; Madaboosi, N.; Albert, J.; Nilsson, M. Padlock Probe Assay for Detection and Subtyping of Seasonal Influenza. Clin. Chem. 2018, 64, 1704–1712.
- Nilsson, M.; Malmgren, H.; Samiotaki, M.; Kwiatkowski, M.; Chowdhary, B.P.; Landegren, U. Padlock probes: Circularizing oligonucleotides for localized DNA detection. Science 1994, 265, 2085–2088.
- Tang, S.M.; Wei, H.; Hu, T.Y.; Jiang, J.Q.; Chang, J.L.; Guan, Y.F.; Zhao, G.J. Suppression of rolling circle amplification by nucleotide analogues in circular template for three DNA polymerases. Biosci. Biotech. Bioch. 2016, 80, 1555–1561.
- Li, D.X.; Zhang, T.T.; Yang, F.; Yuan, R.; Xiang, Y. Efficient and Exponential Rolling Circle Amplification Molecular Network Leads to Ultrasensitive and Label-Free Detection of MicroRNA. Anal. Chem. 2020, 92, 2074–2079.
- Li, X.Y.; Cui, Y.X.; Du, Y.C.; Tang, A.N.; Kong, D.M. Label-Free Telomerase Detection in Single Cell Using a Five-Base Telomerase Product-Triggered Exponential Rolling Circle Amplification Strategy. ACS Sens. 2019, 4, 1090–1096.
- Xu, H.; Xue, C.; Zhang, R.B.; Chen, Y.R.; Li, F.; Shen, Z.F.; Jia, L.; Wu, Z.S. Exponential rolling circle amplification and its sensing application for highly sensitive DNA detection of tumour suppressor gene. Sens. Actuators B Chem. 2017, 243, 1240–1247.
- Pumford, E.A.; Lu, J.; Spaczai, I.; Prasetyo, M.E.; Zheng, E.M.; Zhang, H.; Kamei, D.T. Developments in integrating nucleic acid isothermal amplification and detection systems for point-of-care diagnostics. Biosens. Bioelectron. 2020, 170, 112674.
- Ulanovsky, L.; Bodner, M.; Trifonov, E.N.; Choder, M. Curved DNA: Design, synthesis, and circularization. Proc. Natl. Acad. Sci. USA 1986, 83, 862–866.
- Jin, G.; Wang, C.; Yang, L.; Li, X.; Guo, L.; Qiu, B.; Lin, Z.; Chen, G. Hyperbranched rolling circle amplification based electrochemiluminescence aptasensor for ultrasensitive detection of thrombin. Biosens. Bioelectron. 2015, 63, 166–171.
- Wang, X.M.; Teng, D.; Guan, Q.F.; Tian, F.; Wang, J.H. Detection of genetically modified crops using multiplex asymmetric polymerase chain reaction and asymmetric hyperbranched rolling circle amplification coupled with reverse dot blot. Food Chem. 2015, 173, 1022–1029.
- Yang, L.; Tao, Y.; Yue, G.; Li, R.; Qin, B.; Guo, L.; Lin, Z.; Yang, H.-H. Highly Selective and Sensitive Electrochemiluminescence Biosensor for p53 DNA Sequence Based on Nicking Endonuclease Assisted Target Recycling and Hyperbranched Rolling Circle Amplification. Anal. Chem. 2016, 88, 5097–5103.
- Zhang, L.-R.; Zhu, G.; Zhang, C.-Y. Homogeneous and Label-Free Detection of MicroRNAs Using Bifunctional Strand Displacement Amplification-Mediated Hyperbranched Rolling Circle Amplification. Anal. Chem. 2014, 86, 6703–6709.
- Dahl, F.; Baner, J.; Gullberg, M.; Mendel-Hartvig, M.; Landegren, U.; Nilsson, M. Circle-to-circle amplification for precise and sensitive DNA analysis. Proc. Natl. Acad. Sci. USA 2004, 101, 4548–4553.
- Liu, M.; Yin, Q.X.; McConnell, E.M.; Chang, Y.Y.; Brennan, J.D.; Li, Y.F. DNAzyme Feedback Amplification: Relaying Molecular Recognition to Exponential DNA Amplification. Chem. Eur. J. 2018, 24, 4473–4479.
- Zhao, Y.H.; Wang, Y.; Liu, S.; Wang, C.L.; Liang, J.X.; Li, S.S.; Qu, X.N.; Zhang, R.F.; Yu, J.H.; Huang, J.D. Triple-helix molecular-switch-actuated exponential rolling circular amplification for ultrasensitive fluorescence detection of miRNAs. Analyst 2019, 144, 5245–5253.
- Jiang, Y.; Zou, S.; Cao, X. A simple dendrimer-aptamer based microfluidic platform for E. coli O157:H7 detection and signal intensification by rolling circle amplification. Sens. Actuators B Chem. 2017, 251, 976–984.
- Peng, X.; Liang, W.-B.; Wen, Z.-B.; Xiong, C.-Y.; Zheng, Y.-N.; Chai, Y.-Q.; Yuan, R. Ultrasensitive Fluorescent Assay Based on a Rolling-Circle-Amplification-Assisted Multisite-Strand-Displacement-Reaction Signal-Amplification Strategy. Anal. Chem. 2018, 90, 7474–7479.
- Wang, J.; Dong, H.-Y.; Zhou, Y.; Han, L.-Y.; Zhang, T.; Lin, M.; Wang, C.; Xu, H.; Wu, Z.-S.; Jia, L. Immunomagnetic antibody plus aptamer pseudo-DNA nanocatenane followed by rolling circle amplication for highly sensitive CTC detection. Biosens. Bioelectron. 2018, 122, 239–246.
- Wang, J.; Mao, S.; Li, H.-F.; Lin, J.-M. Multi-DNAzymes-functionalized gold nanoparticles for ultrasensitive chemiluminescence detection of thrombin on microchip. Anal. Chim. Acta 2018, 1027, 76–82.
- Tang, S.R.; Tong, P.; Li, H.; Tang, J.; Zhang, L. Ultrasensitive electrochemical detection of Pb2+ based on rolling circle amplification and quantum dots tagging. Biosens. Bioelectron. 2013, 42, 608–611.
- Cai, W.; Xie, S.B.; Zhang, J.; Tang, D.Y.; Tang, Y. Immobilized-free miniaturized electrochemical sensing system for Pb2+ detection based on dual Pb2+-DNAzyme assistant feedback amplification strategy. Biosens. Bioelectron. 2018, 117, 312–318.
- Cheng, W.; Zhang, W.; Yan, Y.R.; Shen, B.; Zhu, D.; Lei, P.H.; Ding, S.J. A novel electrochemical biosensor for ultrasensitive and specific detection of DNA based on molecular beacon mediated circular strand displacement and rolling circle amplification. Biosens. Bioelectron. 2014, 62, 274–279.
- Mittal, S.; Thakur, S.; Mantha, A.K.; Kaur, H. Bio-analytical applications of nicking endonucleases assisted signal-amplification strategies for detection of cancer biomarkers -DNA methyl transferase and microRNA. Biosens. Bioelectron. 2019, 124, 233–243.
- Bialy, R.M.; Ali, M.M.; Li, Y.F.; Brennan, J.D. Protein-Mediated Suppression of Rolling Circle Amplification for Biosensing with an Aptamer-Containing DNA Primer. Chem. Eur. J. 2020, 26, 5085–5092.
- Fan, T.T.; Mao, Y.; Liu, F.; Zhang, W.; Lin, J.S.; Yin, J.X.; Tan, Y.; Huang, X.T.; Jiang, Y.Y. Label-free fluorescence detection of circulating microRNAs based on duplex-specific nuclease-assisted target recycling coupled with rolling circle amplification. Talanta 2019, 200, 480–486.
- Duy, J.; Smith, R.L.; Collins, S.D.; Connell, L.B. A field-deployable colorimetric bioassay for the rapid and specific detection of ribosomal RNA. Biosens. Bioelectron. 2014, 52, 433–437.
- Wen, Y.Q.; Xu, Y.; Mao, X.H.; Wei, Y.L.; Song, H.Y.; Chen, N.; Huang, Q.; Fan, C.H.; Li, D. DNAzyme-Based Rolling-Circle Amplification DNA Machine for Ultrasensitive Analysis of MicroRNA in Drosophila Larva. Anal. Chem. 2012, 84, 7664–7669.
- Tang, L.H.; Liu, Y.; Ali, M.M.; Kang, D.K.; Zhao, W.A.; Li, J.H. Colorimetric and Ultrasensitive Bioassay Based on a Dual-Amplification System Using Aptamer and DNAzyme. Anal. Chem. 2012, 84, 4711–4717.
- Du, J.; Xu, Q.F.; Lu, X.Q.; Zhang, C.Y. A Label-Free Bioluminescent Sensor for Real-Time Monitoring Polynucleotide Kinase Activity. Anal. Chem. 2014, 86, 8481–8488.
- Mashimo, Y.; Mie, M.; Suzuki, S.; Kobatake, E. Detection of small RNA molecules by a combination of branched rolling circle amplification and bioluminescent pyrophosphate assay. Anal. Bioanal. Chem. 2011, 401, 221–227.




