
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Learn-Han Lee | + 10480 word(s) | 10480 | 2021-09-27 06:12:39 | | | |
| 2 | Vengadesh Letchumanan | + 1 word(s) | 10481 | 2021-10-02 10:40:26 | | | | |
| 3 | Angel Yun-Kuan Thye | -1 word(s) | 10479 | 2021-10-02 11:25:02 | | | | |
| 4 | Priyia Pusparajah | -1 word(s) | 10479 | 2021-10-02 16:16:45 | | | | |
| 5 | Chan Kok Gan | -1 word(s) | 10479 | 2021-10-03 05:07:58 | | | | |
| 6 | Peter Tang | -9 word(s) | 10470 | 2021-10-08 11:18:53 | | |
Video Upload Options
The worldwide battle against the SARS-CoV-2 virus rages on, with millions infected and many innocent lives lost. The causative organism, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is a beta coronavirus that belongs to the Coronaviridae family. Many clinically significant variants have emerged, as the virus’s genome is prone to various mutations, leading to antigenic drift and resulting in evasion of host immune recognition. The current variants of concern (VOCs) include B.1.1.7 (Alpha), B.1.351 (Beta), B.1.617/B.1.617.2 (Delta), and P.1 (Gamma). The emerging variants contain various important mutations on the spike protein, leading to deleterious consequences, such as immune invasion and vaccine escape. These adverse effects result in increased transmissibility, morbidity, and mortality and the evasion of detection by existing or currently available diagnostic tests, potentially delaying diagnosis and treatment. This review discusses the key mutations present in the VOC strains and provides insights into how these mutations allow for greater transmissibility and immune evasion than the progenitor strain. Continuous monitoring and surveillance of VOC strains play a vital role in preventing and controlling the virus’s spread.
1. Introduction
2. Variants of Concern (VOCs)
2.1. B.1.1.7 (Alpha)
2.2. B.1.351 (Beta)
2.3. B.1.617/B.1.617.2 (Delta)
2.4. P.1 (Gamma)
| B.1.1.7 | B.1.351 | B.1.617.2 | P.1 | |
|---|---|---|---|---|
| WHO Label | Alpha [30] | Beta [13] | Delta [62] | Gamma [65] |
| Country First Detected | England [30] | South Africa [13] | India [56] | Brazil [65] |
| First Detected | September 2020 | October 2020 | December 2020 | December 2020 |
| Spike mutations | 69–70HV and 144Y deletions, N501Y, D614G, A570D, P681H, T716I, S982A, D1118H [33] E484K, S494P, and K1191N (found in some sequences) [35] |
L242–244 deletions, A701V, D215G, D80A, D614G, E484K, K417N, N501Y, R246I, L18F [50] | 156–157 deletions, D614G, D950N, L452R, T19R, T478K, P681R, R158GG142D (Found in some) [62] | K417T, E484K, N501Y, L18F, T20N, P26S, D138Y, R190S, D614G, H655Y, V1176F, T1027I [67] |
| Transmissibility | 43–82% more transmissible [30] | 50% more transmissible [53] | 60% more transmissible [61] | Some studies reported 1.7–2.5 times more transmissible [65][74][75] |
| Viral Load | High [23][36] No difference [38] |
High [49] | N/A | High in reinfection case [15] |
| Duration of Infection | Long [26] | N/A | N/A | N/A |
| Hospitalization | High [32][39] | High [32] | High [61] | High [32][74] |
| Mortality | Increase [25][40] | Increase [52] | N/A | Increase [65] |
| Severity | No change [38][42][43] | N/A | N/A | N/A |
| Risk of reinfection | Not higher [42] | High [51][76] | N/A | 6.4% [74] |
| Resistant to antibody neutralization | Resistant to most a mAbs directed against b NTD and slightly resistant to some mAbs directed against the c RBD [44] | Resistant to most mAbs directed against NTD and many mAbs directed against the RBD [44] | N/A | Resistant to some mAbs directed against RBD [69] |
| Resistance against convalescent plasma and sera | Less resistant [11][44] | More resistant [11][44][77] | N/A | Less resistant than B.1.351 [44][78] |
| Vaccine efficacy | Minimal impact [21][22] | Decrease for Pfizer [33], Moderna [79], Novavax, Johnson & Johnson [80][81][82], AstraZeneca [82][83] | 2 doses of Pfizer [84][85][86] or AstraZeneca [84] is still protective | Decrease for CoronaVac [87] |
3. Pathophysiology of SARS-CoV-2 Variants
3.1. Entry of SARS-CoV-2: Spike Glycoprotein/ACE2
3.2. Mutations Affect Binding Affinity (N501Y, E484K/E484Q, K417N/T)
3.3. Mutations Increase Cell Entry and Infectivity (Δ69–70, A570D, S982A, D614G, E484K/Q, K417N/T, P681H/R, L452R)
3.4. Impact of Mutations in The RBD on Plasma Binding and Neutralization (K417N/T, N501Y, E484K/Q, L452R)
3.5. Impact of RBD Mutations on Neutralizing Activity of Convalescent Plasma or Sera (L452R, E484K/Q)
3.6. Impact of Deletions in the NTD (Δ69–70, ΔY144 Deletion, ΔL242–Δ244, and/or R246I)
| Key Mutations | Implications | References |
|---|---|---|
| D614G | Increases human host infectivity and transmission efficiency. | [6][95] |
| Strengthens cleavage efficiency by substituting spike conformational diversity. | [101][124][125] | |
| Δ69–70 deletion | Modifies loop 2 (69–76aa), pulling it nearer to the a NTD. | [29] |
| Increases infectivity by 2-fold over a single round of infection. | [109] | |
| ΔY144 deletion | Loss of binding ability with neutralizing antibodies. | [28][44] |
| ΔL242–Δ244 | Loss of binding ability with neutralizing antibodies. | [28][44] |
| A570D, D614G and S982A | Possibly enhances dynamic viral fusion mechanism via the reduction in intermolecular stability of spike protein subunits. | [34] |
| However, contradicts Hoffman et al., who found that B.1.1.7, B.1.351, and P.1 had no significant difference in spike protein stability and entry kinetics compared with the progenitor isolate with D614G exchange. | [116] | |
| N501Y | Increases binding affinity to b ACE2 due to solid aromatic interactions of π stacking between Y41 (Tyr41) and Y501 (Tyr501), and forming two hydrogen bonds with K353 (Lys353) and D38 (Asp38). | [54][99][105][106][108] |
| Could be the cause for increased transmissibility of B.1.1.7 and B.1.351. | [52][92][109] | |
| Contributes to the escape of some class 1 neutralizing antibodies. | [117][142][143][148] | |
| Antigenic effects limited to a small number of c mAbs, with no significant impact on the neutralizing activity of convalescent plasma or sera from vaccinated individuals. | [44][46][79][149] | |
| Does not drastically affect the overall function of polyclonal T cell responsiveness. | [34] | |
| E484K/Q | Mutation E484K and E484Q have neutral to mildly advantageous effects on the affinity of d RBD for ACE2. | [54] |
| E484K: Favor RBD-up confirmation due to S1 movements opposite of normal E484, which stabilizes the RBD-down confirmation. | [107][111] | |
| In progenitor and B.1.1.7+E484K strains, it disrupts the electrostatic bond, increasing the binding affinity of RBD to ACE2 moderately. | [13][54][112] | |
| In P.1, it forms a strong hydrogen bond with residue E75 (Glu75) on human ACE2, near enough to form a salt bridge, strengthening the binding affinity. | [108] | |
| Results in partial resistance to neutralization. | [44][46][51][78][150][151] | |
| Causes resistance to neutralizing antibodies in class 2 and convalescent sera. | [117][142][143][148] | |
| A few studies found no significant effect on the binding affinity between SARS-CoV- 2 RBD and ACE2. | [108][114] | |
| E484Q: Associated with lower convalescent serum neutralization, neutralization of antibodies, and the ability to reinfect individuals who had not been infected by these mutated variants. | [142][143] | |
| K417N/T | Unfavorable for RBD–ACE2 complex formation. | [108][121] |
| Moderate impact on the binding affinity of RBD–ACE2. | [108] | |
| Escapes neutralization by mAbs. | [71][143][145] | |
| K417N: Destabilizes the RBD-down conformation; increases the tendency for open configuration. | [107] | |
| Stops crucial interactions with class 1 neutralizing antibodies and possibly has a role in immune evasion. | [117][142][143][148] | |
| P681H/R | Causes structural rearrangement and host cell fusion, allowing cell entry. | [34] |
| Contributes to SARS-CoV-2 transmission and infection. | [126][127] | |
| Causes slight ↑ in S1/S2 cleavage, but does not significantly affect viral fitness. | [129] | |
| L452R | Increases infectivity by stabilizing the e S glycoprotein and ACE2 interaction. | [130][131][132] |
| Causes huge increase in free energy at the RBD and ACE2 binding complex, resulting in stronger cell–virus attachment and increased infectivity. | [131][133] | |
| Could evade the human leukocyte antigen (HLA)-24 limited cellular immunity, boost viral infectivity, and possibly stimulate viral replication. | [134] | |
| Can decrease the sensitivity to a few antibodies and human convalescent sera. | [139][158] | |
| N501Y + E484K + K417N | More significant decrease in neutralization compared with any of these mutations alone. | [51][70][71] |
4. Variants of Concern (VOCs) Impact on Vaccine Efficacy
5. Conclusions
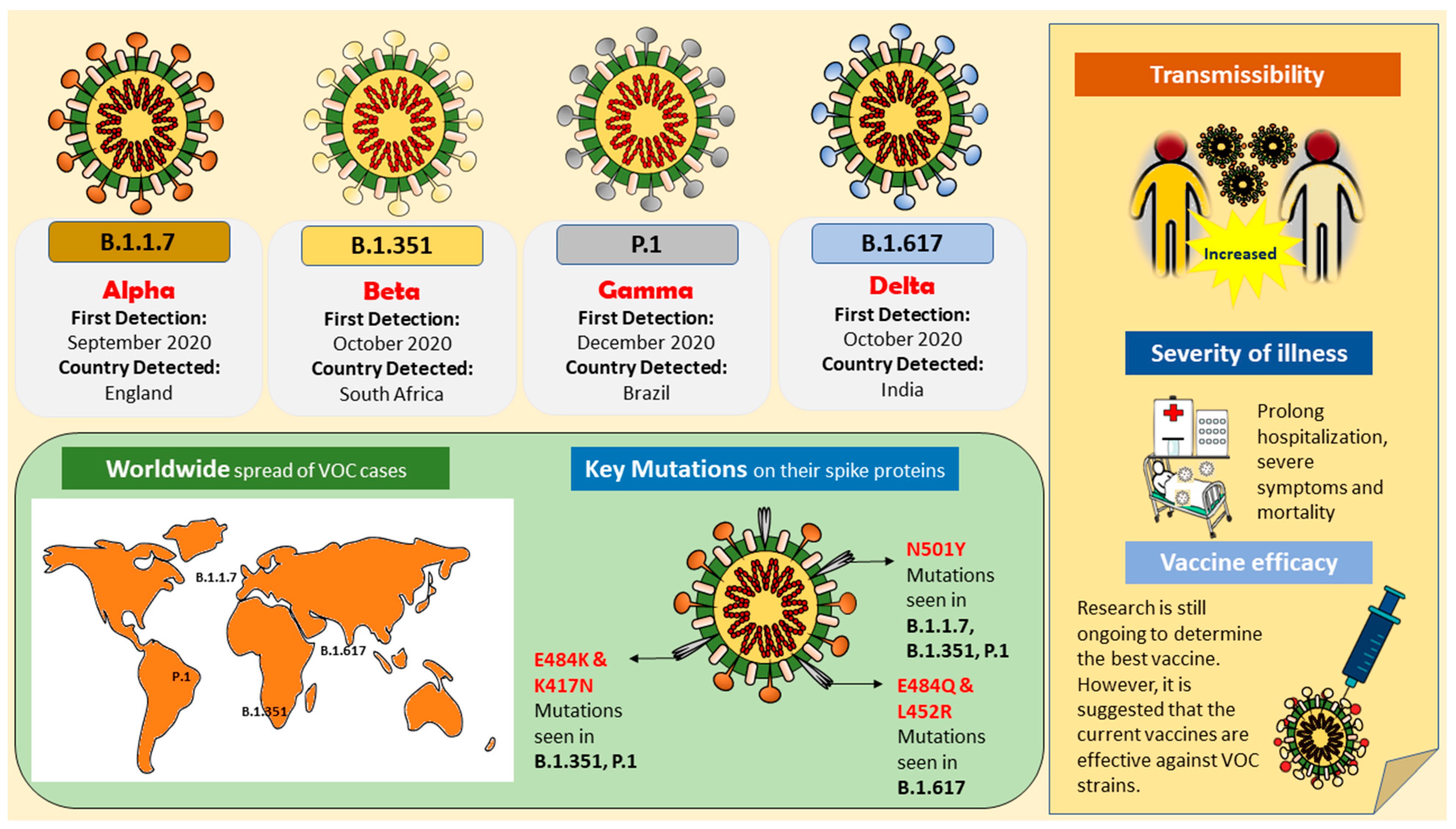 Figure 1. Illustration of variants of concern (VOCs); B.1.1.7 (Alpha), B.1.351 (Beta), B.1.617/B.1.617.2 (Delta), and P.1 (Gamma), their key mutations, and their impact on the public health.
Figure 1. Illustration of variants of concern (VOCs); B.1.1.7 (Alpha), B.1.351 (Beta), B.1.617/B.1.617.2 (Delta), and P.1 (Gamma), their key mutations, and their impact on the public health.References
- Loo, K.-Y.; Letchumanan, V.; Ser, H.-L.; Teoh, S.L.; Law, J.W.-F.; Tan, L.T.-H.; Ab Mutalib, N.-S.; Chan, K.-G.; Lee, L.-H. COVID-19: Insights into potential vaccines. Microorganisms 2021, 9, 605.
- Johnson, D.; Ren, S.E.C.; Johnson, H.D.; Letchumanan, V. COVID-19: Are Malaysians embracing or suffering the new normality? Prog. Microbes Mol. Biol. 2020, 3.
- Letchumanan, V.; Ab Mutalib, N.-S.; Goh, B.-H.; Lee, L.-H. Novel coronavirus 2019-nCoV: Could this virus become a possible global pandemic. Prog. Microbes Mol. Biol. 2020, 3.
- Lee, V.S.; Chong, W.L.; Sukumaran, S.D.; Nimmanpipug, P.; Letchumanan, V.; Goh, B.H.; Lee, L.-H.; Zain, S.M.; Abd Rahman, N. Computational screening and identifying binding interaction of anti-viral and anti-malarial drugs: Toward the potential cure for SARS-CoV-2. Prog. Drug Discov. Biomed. Sci. 2020, 3.
- Loh, H.C.; Seah, Y.K.; Looi, I. The COVID-19 pandemic and diet change. Prog. Microbes Mol. Biol. 2021, 4.
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B. Tracking changes in SARS-CoV-2 Spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020, 182, 812–827.e819.
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273.
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733.
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292.e286.
- Lauring, A.S.; Hodcroft, E.B. Genetic Variants of SARS-CoV-2—What Do They Mean? JAMA 2021, 325, 529–531.
- Grubaugh, N.D.; Petrone, M.E.; Holmes, E.C. We shouldn’t worry when a virus mutates during disease outbreaks. Nat. Microbiol. 2020, 5, 529–530.
- Tang, J.W.; Tambyah, P.A.; Hui, D.S. Emergence of a new SARS-CoV-2 variant in the UK. J. Infect. 2020, 82, e27–e28.
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020.
- Samarasekera, U. India grapples with second wave of COVID-19. Lancet Microbe 2021, 2, e238.
- Naveca, F.; da Costa, C.; Nascimento, V.; Souza, V.; Corado, A.; Nascimento, F.; Costa, Á.; Duarte, D.; Silva, G.; Mejía, M. SARS-CoV-2 reinfection by the new Variant of Concern (VOC) P. 1 in Amazonas, Brazil. Virological. Org. 2021. Available online: https://virological.org/t/sars-cov-2-reinfection-by-the-new-variant-of-concern-voc-p-1-in-amazonas-brazil/596 (accessed on 7 June 2021).
- Krammer, F. SARS-CoV-2 vaccines in development. Nature 2020, 586, 516–527.
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020, 383, 2603–2615.
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E. Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: A pooled analysis of four randomised trials. Lancet 2021, 397, 881–891.
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021, 384, 403–416.
- Loo, K.-Y.; Letchumanan, V. COVID-19: Malaysia’s fight against this deadly virus. Prog. Microbes Mol. Biol. 2021, 4.
- Shen, X.; Tang, H.; McDanal, C.; Wagh, K.; Fischer, W.; Theiler, J.; Yoon, H.; Li, D.; Haynes, B.F.; Sanders, K.O. SARS-CoV-2 variant B. 1.1. 7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe 2021, 29, 529–539.e523.
- Edara, V.V.; Floyd, K.; Lai, L.; Gardner, M.; Hudson, W.; Piantadosi, A.; Waggoner, J.; Babiker, A.; Ahmed, R.; Xie, X. Infection and mRNA-1273 vaccine antibodies neutralize SARS-CoV-2 UK variant. medRxiv 2021.
- Calistri, P.; Amato, L.; Puglia, I.; Cito, F.; Di Giuseppe, A.; Danzetta, M.L.; Morelli, D.; Di Domenico, M.; Caporale, M.; Scialabba, S. Infection sustained by lineage B. 1.1. 7 of SARS-CoV-2 is characterised by longer persistence and higher viral RNA loads in nasopharyngeal swabs. Int. J. Infect. Dis. 2021, 105, 753–755.
- Davies, N.G.; Jarvis, C.I.; Edmunds, W.J.; Jewell, N.P.; Diaz-Ordaz, K.; Keogh, R.H. Increased mortality in community-tested cases of SARS-CoV-2 lineage B. 1.1. 7. Nature 2021, 593, 270–274.
- Grint, D.J.; Wing, K.; Williamson, E.; McDonald, H.I.; Bhaskaran, K.; Evans, D.; Evans, S.J.; Walker, A.J.; Hickman, G.; Nightingale, E. Case fatality risk of the SARS-CoV-2 variant of concern B. 1.1. 7 in England, 16 November to 5 February. Euro Surveill. 2021, 26, 2100256.
- Kissler, S.M.; Fauver, J.R.; Mack, C.; Tai, C.; Breban, M.; Watkins, A.E.; Samant, R.; Anderson, D.; Ho, D.; Grubaugh, N.D. Densely sampled viral trajectories suggest longer duration of acute infection with B. 1.1. 7 variant relative to non-B. 1.1. 7 SARS-CoV-2. medRxiv 2021.
- Choi, B.; Choudhary, M.C.; Regan, J.; Sparks, J.A.; Padera, R.F.; Qiu, X.; Solomon, I.H.; Kuo, H.-H.; Boucau, J.; Bowman, K. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. J. Med. 2020, 383, 2291–2293.
- McCarthy, K.R.; Rennick, L.J.; Nambulli, S.; Robinson-McCarthy, L.R.; Bain, W.G.; Haidar, G.; Duprex, W.P. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science 2021, 371, 1139–1142.
- Kemp, S.A.; Collier, D.A.; Datir, R.P.; Ferreira, I.A.; Gayed, S.; Jahun, A.; Hosmillo, M.; Rees-Spear, C.; Mlcochova, P.; Lumb, I.U. SARS-CoV-2 evolution during treatment of chronic infection. Nature 2021, 592, 277–282.
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.; Russell, T.W.; Tully, D.C.; Washburne, A.D. Estimated transmissibility and impact of SARS-CoV-2 lineage B. 1.1. 7 in England. Science 2021, 372, 6538.
- Courjon, J.-V.; Contenti, J.; Demonchy, E.; Levraut, J.; Barbry, P.; Rios, G.; Dellamonica, J.; Chirio, D.; Bonnefoy, C.; Giordanengo, V. Spread of the SARS-CoV-2 UK-variant in the South East of France: Impact on COVID-19 patients age, comorbidity profiles and clinical presentation, week 50 2020 to week 8 2021. medRxiv 2021.
- Funk, T.; Pharris, A.; Spiteri, G.; Bundle, N.; Melidou, A.; Carr, M.; Gonzalez, G.; Garcia-Leon, A.; Crispie, F.; O’Connor, L. Characteristics of SARS-CoV-2 variants of concern B. 1.1. 7, B. 1.351 or P. 1: Data from seven EU/EEA countries, weeks 38/2020 to 10/2021. Euro Surveill. 2021, 26, 2100348.
- Supasa, P.; Zhou, D.; Dejnirattisai, W.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.; Nutalai, R.; Tuekprakhon, A. Reduced neutralization of SARS-CoV-2 B. 1.1. 7 variant by convalescent and vaccine sea. Cell 2021, 184, 2201–2211.
- Ostrov, D.A. Structural Consequences of Variation in SARS-CoV-2 B. 1.1. 7. J. Cell. Immunol. 2021, 3, 103.
- SARS-CoV-2 Variant Classifications and Definitions. Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html (accessed on 24 June 2021).
- Kidd, M.; Richter, A.; Best, A.; Cumley, N.; Mirza, J.; Percival, B.; Mayhew, M.; Megram, O.; Ashford, F.; White, T. S-Variant SARS-CoV-2 Lineage B1. 1.7 Is Associated With Significantly Higher Viral Load in Samples Tested by TaqPath Polymerase Chain Reaction. J. Infect. Dis. 2021, 223, 1666–1670.
- Team, E.E. Updated rapid risk assessment from ECDC on the risk related to the spread of new SARS-CoV-2 variants of concern in the EU/EEA–first update. Eurosurveillance 2021, 26, 2101211.
- Walker, A.S.; Vihta, K.D.; Gethings, O.; Pritchard, E.; Jones, J.; House, T.; Bell, I.; Bell, J.; Newton, J.; Farrar, J. Increased infections, but not viral burden, with a new SARS-CoV-2 variant. medRxiv 2021.
- Bager, P.; Wohlfahrt, J.; Fonager, J.; Albertsen, M.; Yssing Michaelsen, T.; Holten Møller, C.; Ethelberg, S.; Legarth, R.; Fischer Button, M.S.; Gubbels, S.M. Increased risk of hospitalisation associated with infection with SARS-CoV-2 lineage B. 1.1. 7 in Denmark. SSRN Electron. J. 2021.
- Challen, R.; Brooks-Pollock, E.; Read, J.M.; Dyson, L.; Tsaneva-Atanasova, K.; Danon, L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: Matched cohort study. BMJ 2021, 372.
- Horby, P.H.C.; Davies, N.; Edmunds, J.; Ferguson, N.; Medley, G.; Semple, C. NERVTAG Note on B. 1.1. 7 Severity. Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/961037/NERVTAG_note_on_B.1.1.7_severity_for_SAGE_77__1_.pdf (accessed on 7 June 2021).
- Graham, M.S.; Sudre, C.H.; May, A.; Antonelli, M.; Murray, B.; Varsavsky, T.; Kläser, K.; Canas, L.S.; Molteni, E.; Modat, M. Changes in symptomatology, reinfection, and transmissibility associated with the SARS-CoV-2 variant B. 1.1. 7: An ecological study. Lancet Public Health 2021, 6, e335–e345.
- Frampton, D.; Rampling, T.; Cross, A.; Bailey, H.; Heaney, J.; Byott, M.; Scott, R.; Sconza, R.; Price, J.; Margaritis, M. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B. 1.1. 7 lineage in London, UK: A whole-genome sequencing and hospital-based cohort study. Lancet Infect. Dis. 2021, 21, 1246–1256.
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D. Antibody resistance of SARS-CoV-2 variants B. 1.351 and B. 1.1. 7. Nature 2021, 593, 130–135.
- Rathnasinghe, R.; Jangra, S.; Cupic, A.; Martínez-Romero, C.; Mulder, L.C.; Kehrer, T.; Yildiz, S.; Choi, A.; Mena, I.; De Vrieze, J. The N501Y mutation in SARS-CoV-2 spike leads to morbidity in obese and aged mice and is neutralized by convalescent and post-vaccination human sera. MedRxiv 2021.
- Xie, X.; Liu, Y.; Liu, J.; Zhang, X.; Zou, J.; Fontes-Garfias, C.R.; Xia, H.; Swanson, K.A.; Cutler, M.; Cooper, D. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021, 27, 620–621.
- Emary, K.; Golubchik, T.; Aley, P.K.; Ariani, C.V.; Angus, B.; Bibi, S.; Blane, B.; Bonsall, D.; Cicconi, P.; Charlton, S.; et al. Oxford COVID-19 Vaccine Trial Group (2021). Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): An exploratory analysis of a randomised controlled trial. Lancet 2021, 397, 1351–1362.
- Tang, J.W.; Toovey, O.T.; Harvey, K.N.; Hui, D.D. Introduction of the South African SARS-CoV-2 variant 501Y. V2 into the UK. J. Infect. 2021, 82, e8–e10.
- Boehm, E.; Kronig, I.; Neher, R.A.; Eckerle, I.; Vetter, P.; Kaiser, L. Novel SARS-CoV-2 variants: The pandemics within the pandemic. Clin. Microbiol. Infect. 2021, 27, 1109–1117.
- Tegally, H.; Wilkinson, E.; Lessells, R.J.; Giandhari, J.; Pillay, S.; Msomi, N.; Mlisana, K.; Bhiman, J.N.; von Gottberg, A.; Walaza, S. Sixteen novel lineages of SARS-CoV-2 in South Africa. Nat. Med. 2021, 27, 440–446.
- Wibmer, C.K.; Ayres, F.; Hermanus, T.; Madzivhandila, M.; Kgagudi, P.; Oosthuysen, B.; Lambson, B.E.; De Oliveira, T.; Vermeulen, M.; Van der Berg, K. SARS-CoV-2 501Y. V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021, 27, 622–625.
- Abdool Karim, S.S.; de Oliveira, T. New SARS-CoV-2 variants—Clinical, public health, and vaccine implications. N. Engl. J. Med. 2021, 384, 1866–1868.
- van Oosterhout, C.; Hall, N.; Ly, H.; Tyler, K.M. COVID-19 evolution during the pandemic–Implications of new SARS-CoV-2 variants on disease control and public health policies. Virulence 2021, 12, 507–508.
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 2020, 182, 1295–1310.e1220.
- Faria, N.R.; Claro, I.M.; Candido, D.; Franco, L.M.; Andrade, P.S.; Coletti, T.M.; Silva, C.A.; Sales, F.C.; Manuli, E.R.; Aguiar, R.S. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: Preliminary findings. Virological 2021, 372, 815–821.
- Threat Assessment Brief: Emergence of SARS-CoV-2 B.1.617 Variants in India and Situation in the EU/EEA. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/Emergence-of-SARS-CoV-2-B.1.617-variants-in-India-and-situation-in-the-EUEEA_0.pdf (accessed on 8 June 2021).
- Shan, L.; Agarwal, V. What We Know About India’s Covid-19 Variant B.1.617. Available online: https://www.wsj.com/articles/what-we-know-about-indias-double-mutant-covid-19-variant-11619193481 (accessed on 16 May 2021).
- Khaitan, S. Most Children Getting Mild COVID-19, Even Severe Cases Treatable. Available online: https://www.indiaspend.com/covid-19/most-children-getting-mild-covid-19-even-severe-cases-treatable-746129 (accessed on 7 June 2021).
- Covid, C.; Team, R.; Covid, C.; Team, R.; COVID, C.; Team, R.; Bialek, S.; Gierke, R.; Hughes, M.; McNamara, L.A. Coronavirus disease 2019 in children—United States, February 12–April 2, 2020. Morb. Mortal. Wkly. Rep. 2020, 69, 422.
- Kim, L.; Whitaker, M.; O’Halloran, A.; Kambhampati, A.; Chai, S.J.; Reingold, A.; Armistead, I.; Kawasaki, B.; Meek, J.; Yousey-Hindes, K. Hospitalization rates and characteristics of children aged <18 years hospitalized with laboratory-confirmed COVID-19—COVID-NET, 14 States, March 1–July 25, 2020. Morb. Mortal. Wkly. Rep. 2020, 69, 1081.
- Mahase, E. Delta variant: What is happening with transmission, hospital admissions, and restrictions? BMJ 2021.
- Srivastava, S.; Banu, S.; Singh, P.; Sowpati, D.T.; Mishra, R.K. SARS-CoV-2 genomics: An Indian perspective on sequencing viral variants. J. Biosci. 2021, 46, 1–14.
- Yadav, P.D.; Mohandas, S.; Shete, A.M.; Nyayanit, D.A.; Gupta, N.; Patil, D.Y.; Sapkal, G.N.; Potdar, V.; Kadam, M.; Kumar, S. SARS CoV-2 variant B. 1.617. 1 is highly pathogenic in hamsters than B. 1 variant. bioRxiv 2021.
- Adam, D. What scientists know about new, fast-spreading coronavirus variants. Nature 2021, 594, 19–20.
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; Candido, D.d.S.; Mishra, S.; Crispim, M.A.; Sales, F.C.; Hawryluk, I.; McCrone, J.T. Genomics and epidemiology of the P. 1 SARS-CoV-2 lineage in Manaus, Brazil. Science 2021, 372, 815–821.
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407.
- Dejnirattisai, W.; Zhou, D.; Supasa, P.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.; Tuekprakhon, A.; Nutalai, R. Antibody evasion by the P. 1 strain of SARS-CoV-2. Cell 2021, 184, 2939–2954.e2939.
- Fujino, T.; Nomoto, H.; Kutsuna, S.; Ujiie, M.; Suzuki, T.; Sato, R.; Fujimoto, T.; Kuroda, M.; Wakita, T.; Ohmagari, N. Novel SARS-CoV-2 variant in travelers from Brazil to Japan. Emerg. Infect. Dis. 2021, 27, 1243.
- Wang, P.; Casner, R.G.; Nair, M.S.; Wang, M.; Yu, J.; Cerutti, G.; Liu, L.; Kwong, P.D.; Huang, Y.; Shapiro, L. Increased resistance of SARS-CoV-2 variant P. 1 to antibody neutralization. Cell Host Microbe 2021, 29, 747–751.e744.
- Cele, S.; Gazy, I.; Jackson, L.; Hwa, S.-H.; Tegally, H.; Lustig, G.; Giandhari, J.; Pillay, S.; Wilkinson, E.; Naidoo, Y. Escape of SARS-CoV-2 501Y. V2 from neutralization by convalescent plasma. Nature 2021, 593, 142–146.
- Wang, Z.; Schmidt, F.; Weisblum, Y.; Muecksch, F.; Barnes, C.O.; Finkin, S.; Schaefer-Babajew, D.; Cipolla, M.; Gaebler, C.; Lieberman, J.A. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 2021, 592, 616–622.
- Greaney, A.J.; Loes, A.N.; Crawford, K.H.; Starr, T.N.; Malone, K.D.; Chu, H.Y.; Bloom, J.D. Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. Cell Host Microbe 2021, 29, 463–476.
- Sabino, E.C.; Buss, L.F.; Carvalho, M.P.; Prete, C.A.; Crispim, M.A.; Fraiji, N.A.; Pereira, R.H.; Parag, K.V.; da Silva Peixoto, P.; Kraemer, M.U. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet 2021, 397, 452–455.
- Coutinho, R.M.; Marquitti, F.M.D.; Ferreira, L.S.; Borges, M.E.; da Silva, R.L.P.; Canton, O.; Portella, T.P.; Lyra, S.P.; Franco, C.; da Silva, A.A.M. Model-based evaluation of transmissibility and reinfection for the P. 1 variant of the SARS-CoV-2. medRxiv 2021.
- Naveca, F.; Nascimento, V.; Souza, V.; Corado, A.; Nascimiento, F.; Silva, G.; Costa, A.; Duarte, D.; Pessoa, K.; Mejia, M.; et al. COVID-19 epidemic in the Brazilian state of Amazonas was driven by long-term persistence of endemic SARS-CoV2 lineages and the recent emergence of the new Variant of Concern, P.1. Res. Sq. 2021. Available online: https://www.researchsquare.com/article/rs-275494/v1 (accessed on 17 June 2021).
- Zucman, N.; Uhel, F.; Descamps, D.; Roux, D.; Ricard, J.-D. Severe Reinfection with South African Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Variant 501Y. V2. Clin. Infect. Dis. 2021.
- Zhou, D.; Dejnirattisai, W.; Supasa, P.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.; Tuekprakhon, A.; Nutalai, R. Evidence of escape of SARS-CoV-2 variant B. 1.351 from natural and vaccine-induced sera. Cell 2021, 184, 2348–2361.e2346.
- Garcia-Beltran, W.F.; Lam, E.C.; Denis, K.S.; Nitido, A.D.; Garcia, Z.H.; Hauser, B.M.; Feldman, J.; Pavlovic, M.N.; Gregory, D.J.; Poznansky, M.C. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 2021, 184, 2372–2383.e2379.
- Wu, K.; Werner, A.P.; Moliva, J.I.; Koch, M.; Choi, A.; Stewart-Jones, G.B.; Bennett, H.; Boyoglu-Barnum, S.; Shi, W.; Graham, B.S. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv 2021.
- Callaway, E.; Mallapaty, S. Novavx covid vaccine protects people against variants. Nature 2021, 590, 17.
- Wadman, M.; Cohen, J. Novavax vaccine delivers 89% efficacy against COVID-19 in UK—But is less potent in South Africa. Science 2021.
- Dagan, N.; Barda, N.; Kepten, E.; Miron, O.; Perchik, S.; Katz, M.A.; Hernán, M.A.; Lipsitch, M.; Reis, B.; Balicer, R.D. BNT162b2 mRNA Covid-19 vaccine in a nationwide mass vaccination setting. N. Engl. J. Med. 2021, 384, 1412–1423.
- Madhi, S.A.; Baillie, V.; Cutland, C.L.; Voysey, M.; Koen, A.L.; Fairlie, L.; Padayachee, S.D.; Dheda, K.; Barnabas, S.L.; Bhorat, Q.E. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B. 1.351 variant. N. Engl. J. Med. 2021, 384, 1885–1898.
- Vaccines Highly Effective Against B.1.617.2 Variant after 2 Doses . Available online: https://www.gov.uk/government/news/vaccines-highly-effective-against-b-1-617-2-variant-after-2-doses (accessed on 27 June 2021).
- Wall, E.C.; Wu, M.; Harvey, R.; Kelly, G.; Warchal, S.; Sawyer, C.; Daniels, R.; Hobson, P.; Hatipoglu, E.; Ngai, Y. Neutralising antibody activity against SARS-CoV-2 VOCs B. 1.617. 2 and B. 1.351 by BNT162b2 vaccination. Lancet 2021, 397, 2331–2333.
- Liu, J.; Liu, Y.; Xia, H.; Zou, J.; Weaver, S.C.; Swanson, K.A.; Cai, H.; Cutler, M.; Cooper, D.; Muik, A. BNT162b2-elicited neutralization of B. 1.617 and other SARS-CoV-2 variants. Nature 2021, 596, 273–275.
- Cohen, J.; Moutinho, S. Third time’s the charm? Brazil scales back efficacy claims for COVID-19 vaccine from China. Science 2021, 1126.
- Ribes, M.; Chaccour, C.; Moncunill, G. Adapt or perish: SARS-CoV-2 antibody escape variants defined by deletions in the Spike N-terminal Domain. Signal Transduct. Target. Ther. 2021, 6, 1–2.
- Gribble, J.; Stevens, L.J.; Agostini, M.L.; Anderson-Daniels, J.; Chappell, J.D.; Lu, X.; Pruijssers, A.J.; Routh, A.L.; Denison, M.R. The coronavirus proofreading exoribonuclease mediates extensive viral recombination. Plos Path. 2021, 17, e1009226.
- Ser, H.-L.; Tan, L.T.-H.; Law, J.W.-F.; Letchumanan, V.; Ab Mutalib, N.-S.; Lee, L.-H. Genomic analysis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) strains isolated in Malaysia. Prog. Microbes Mol. Biol. 2020, 3.
- Science Brief: Emerging SARS-CoV-2 Variants. Available online: https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-emerging-variants.html (accessed on 20 May 2021).
- Rambaut, A.; Loman, N.; Pybus, O.; Barclay, W.; Barrett, J.; Carabelli, A.; Connor, T.; Peacock, T.; Robertson, D.L.; Volz, E. Preliminary Genomic Characterisation of an Emergent SARS-CoV-2 Lineage in the UK Defined by a Novel Set of Spike Mutations. Available online: https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 (accessed on 7 June 2021).
- Hirotsu, Y.; Omata, M. Discovery of a SARS-CoV-2 variant from the P. 1 lineage harboring K417T/E484K/N501Y mutations in Kofu, Japan. J. Infect. 2021, 82, 276–316.
- McCallum, M.; De Marco, A.; Lempp, F.; Tortorici, M.; Pinto, D.; Walls, A. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 2021, 184, 2332–2347.
- Volz, E.; Hill, V.; McCrone, J.T.; Price, A.; Jorgensen, D.; O’Toole, Á.; Southgate, J.; Johnson, R.; Jackson, B.; Nascimento, F.F. Evaluating the effects of SARS-CoV-2 Spike mutation D614G on transmissibility and pathogenicity. Cell 2021, 184, 64–75.e11.
- Leung, K.; Shum, M.H.; Leung, G.M.; Lam, T.T.; Wu, J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Eurosurveillance 2021, 26, 2002106.
- Bar-On, Y.M.; Flamholz, A.; Phillips, R.; Milo, R. Science Forum: SARS-CoV-2 (COVID-19) by the numbers. Elife 2020, 9, e57309.
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e278.
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224.
- Hikmet, F.; Méar, L.; Edvinsson, Å.; Micke, P.; Uhlén, M.; Lindskog, C. The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 2020, 16, e9610.
- Kim, Y.J.; Jang, U.S.; Soh, S.M.; Lee, J.-Y.; Lee, H.-R. The Impact on Infectivity and Neutralization Efficiency of SARS-CoV-2 Lineage B. 1.351 Pseudovirus. Viruses 2021, 13, 633.
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020, 94, e00127-20.
- Cerutti, G.; Guo, Y.; Zhou, T.; Gorman, J.; Lee, M.; Rapp, M.; Reddem, E.R.; Yu, J.; Bahna, F.; Bimela, J. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe 2021, 29, 819–833.e817.
- Yang, L.; Liu, W.; Yu, X.; Wu, M.; Reichert, J.M.; Ho, M. COVID-19 antibody therapeutics tracker: A global online database of antibody therapeutics for the prevention and treatment of COVID-19. Antib. Ther. 2020, 3, 205–212.
- Liu, H.; Zhang, Q.; Wei, P.; Chen, Z.; Aviszus, K.; Yang, J.; Downing, W.; Jiang, C.; Liang, B.; Reynoso, L. The basis of a more contagious 501Y. V1 variant of SARS-COV-2. Cell Res. 2021, 31, 720–722.
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220.
- Gobeil, S.; Janowska, K.; McDowell, S.; Mansouri, K.; Parks, R.; Stalls, V.; Kopp, M.F.; Manne, K.; Saunders, K.O.N.; Edwards, R.J. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation and antigenicity. bioRxiv 2021.
- Shahhosseini, N.; Babuadze, G.G.; Wong, G.; Kobinger, G.P. Mutation Signatures and In Silico Docking of Novel SARS-CoV-2 Variants of Concern. Microorganisms 2021, 9, 926.
- Kemp, S.; Harvey, W.; Datir, R.; Collier, D.; Ferreira, I.; Carabelii, A.; Robertson, D.L.; Gupta, R.K. Recurrent emergence and transmission of a SARS-CoV-2 Spike deletion ΔH69/V70. bioRxiv 2020.
- Kupferschmidt, K. New mutations raise specter of ‘immune escape’. Science 2021, 371, 329–330.
- Winger, A.; Caspari, T. The Spike of Concern—The Novel Variants of SARS-CoV-2. Viruses 2021, 13, 1002.
- Cherian, S.; Potdar, V.; Jadhav, S.; Yadav, P.; Gupta, N.; Das, M.; Rakshit, P.; Singh, S.; Abraham, P.; Panda, S. Convergent evolution of SARS-CoV-2 spike mutations, L452R, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. bioRxiv 2021.
- Tada, T.; Zhou, H.; Dcosta, B.M.; Samanovic, M.I.; Mulligan, M.J.; Landau, N.R. The Spike Proteins of SARS-CoV-2 B. 1.617 and B. 1.618 Variants Identified in India Provide Partial Resistance to Vaccine-elicited and Therapeutic Monoclonal Antibodies. BioRxiv 2021.
- Liu, H.; Wei, P.; Zhang, Q.; Chen, Z.; Aviszus, K.; Downing, W.; Peterson, S.; Reynoso, L.; Downey, G.P.; Frankel, S.K. 501Y. V2 and 501Y. V3 variants of SARS-CoV-2 lose binding to Bamlanivimab in vitro. In Proceedings of the MAbs; Taylor & Francis: Abingdon, UK, 2021; p. 1919285.
- Nelson, G.; Buzko, O.; Spilman, P.R.; Niazi, K.; Rabizadeh, S.; Soon-Shiong, P.R. Molecular dynamic simulation reveals E484K mutation enhances spike RBD-ACE2 affinity and the combination of E484K, K417N and N501Y mutations (501Y. V2 variant) induces conformational change greater than N501Y mutant alone, potentially resulting in an escape mutant. BioRxiv 2021.
- Hoffmann, M.; Arora, P.; Groß, R.; Seidel, A.; Hörnich, B.F.; Hahn, A.S.; Krüger, N.; Graichen, L.; Hofmann-Winkler, H.; Kempf, A. SARS-CoV-2 variants B. 1.351 and P. 1 escape from neutralizing antibodies. Cell 2021, 184, 2384–2393.e2312.
- Weisblum, Y.; Schmidt, F.; Zhang, F.; DaSilva, J.; Poston, D.; Lorenzi, J.C.; Muecksch, F.; Rutkowska, M.; Hoffmann, H.-H.; Michailidis, E. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife 2020, 9, e61312.
- Wang, Y.; Liu, M.; Gao, J. Enhanced receptor binding of SARS-CoV-2 through networks of hydrogen-bonding and hydrophobic interactions. Proc. Natl. Acad. Sci. USA 2020, 117, 13967–13974.
- Yi, C.; Sun, X.; Ye, J.; Ding, L.; Liu, M.; Yang, Z.; Lu, X.; Zhang, Y.; Ma, L.; Gu, W. Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell. Mol. Immunol. 2020, 17, 621–630.
- Ghorbani, M.; Brooks, B.R.; Klauda, J.B. Critical Sequence Hotspots for Binding of Novel Coronavirus to Angiotensin Converter Enzyme as Evaluated by Molecular Simulations. J. Phys. Chem. 2020, 124, 10034–10047.
- Villoutreix, B.O.; Calvez, V.; Marcelin, A.-G.; Khatib, A.-M. In silico investigation of the new UK (B. 1.1. 7) and South African (501y. v2) SARS-CoV-2 variants with a focus at the ace2–spike rbd interface. Int. J. Mol. Sci. 2021, 22, 1695.
- Fratev, F. The N501Y and K417N mutations in the spike protein of SARS-CoV-2 alter the interactions with both hACE2 and human derived antibody: A Free energy of perturbation study. bioRxiv 2020.
- Buchrieser, J.; Dufloo, J.; Hubert, M.; Monel, B.; Planas, D.; Rajah, M.M.; Planchais, C.; Porrot, F.; Guivel-Benhassine, F.; Van der Werf, S. Syncytia formation by SARS-CoV-2-infected cells. Embo J. 2021, 40, e107405.
- Gobeil, S.M.-C.; Janowska, K.; McDowell, S.; Mansouri, K.; Parks, R.; Manne, K.; Stalls, V.; Kopp, M.F.; Henderson, R.; Edwards, R.J. D614G mutation alters SARS-CoV-2 spike conformation and enhances protease cleavage at the S1/S2 junction. Cell Rep. 2021, 34, 108630.
- Yadav, P.; Sapkal, G.N.; Abraham, P.; Ella, R.; Deshpande, G.; Patil, D.Y.; Nyayanit, D.; Gupta, N.; Sahay, R.R.; Shete, A.M. Neutralization of variant under investigation B. 1.617 with sera of BBV152 vaccinees. bioRxiv 2021.
- Hoffmann, M.; Kleine-Weber, H.; Pöhlmann, S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell 2020, 78, 779–784.e775.
- Peacock, T.P.; Goldhill, D.H.; Zhou, J.; Baillon, L.; Frise, R.; Swann, O.C.; Kugathasan, R.; Penn, R.; Brown, J.C.; Sanchez-David, R.Y. The furin cleavage site of SARS-CoV-2 spike protein is a key determinant for transmission due to enhanced replication in airway cells. bioRxiv 2020.
- Zhu, Y.; Feng, F.; Hu, G.; Wang, Y.; Yu, Y.; Zhu, Y.; Xu, W.; Cai, X.; Sun, Z.; Han, W. The S1/S2 boundary of SARS-CoV-2 spike protein modulates cell entry pathways and transmission. BioRxiv 2020.
- Lubinski, B.; Tang, T.; Daniel, S.; Jaimes, J.A.; Whittaker, G. Functional evaluation of proteolytic activation for the SARS-CoV-2 variant B. 1.1. 7: Role of the P681H mutation. bioRxiv 2021.
- Teng, S.; Sobitan, A.; Rhoades, R.; Liu, D.; Tang, Q. Systemic effects of missense mutations on SARS-CoV-2 spike glycoprotein stability and receptor-binding affinity. Brief. Bioinform. 2021, 22, 1239–1253.
- Chen, J.; Wang, R.; Wang, M.; Wei, G.-W. Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol. 2020, 432, 5212–5226.
- Deng, X.; Garcia-Knight, M.A.; Khalid, M.M.; Servellita, V.; Wang, C.; Morris, M.K.; Sotomayor-González, A.; Glasner, D.R.; Reyes, K.R.; Gliwa, A.S. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell 2021, 184, 3426–3437.e8.
- Tchesnokova, V.; Kulakesara, H.; Larson, L.; Bowers, V.; Rechkina, E.; Kisiela, D.; Sledneva, Y.; Choudhury, D.; Maslova, I.; Deng, K. Acquisition of the L452R mutation in the ACE2-binding interface of Spike protein triggers recent massive expansion of SARS-Cov-2 variants. bioRxiv 2021.
- Motozono, C.; Toyoda, M.; Zahradnik, J.; Ikeda, T.; Saito, A.; Tan, T.S.; Ngare, I.; Nasser, H.; Kimura, I.; Uriu, K. An emerging SARS-CoV-2 mutant evading cellular immunity and increasing viral infectivity. BioRxiv 2021.
- Voss, W.N.; Hou, Y.J.; Johnson, N.V.; Kim, J.E.; Delidakis, G.; Horton, A.P.; Bartzoka, F.; Paresi, C.J.; Tanno, Y.; Abbasi, S.A. Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes in COVID-19 convalescent plasma. bioRxiv 2020.
- Seydoux, E.; Homad, L.J.; MacCamy, A.J.; Parks, K.R.; Hurlburt, N.K.; Jennewein, M.F.; Akins, N.R.; Stuart, A.B.; Wan, Y.-H.; Feng, J. Analysis of a SARS-CoV-2-infected individual reveals development of potent neutralizing antibodies with limited somatic mutation. Immunity 2020, 53, 98–105.e105.
- Brouwer, P.J.; Caniels, T.G.; van der Straten, K.; Snitselaar, J.L.; Aldon, Y.; Bangaru, S.; Torres, J.L.; Okba, N.M.; Claireaux, M.; Kerster, G. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 2020, 369, 643–650.
- Huang, K.-Y.A.; Tan, T.; Chen, T.-H.; Huang, C.-G.; Harvey, R.; Hussain, S.; Chen, C.-P.; Harding, A.; Gilbert-Jaramillo, J.; Liu, X. Plasmablast-derived antibody response to acute SARS-CoV-2 infection in humans. bioRxiv 2020.
- Li, Q.; Wu, J.; Nie, J.; Zhang, L.; Hao, H.; Liu, S.; Zhao, C.; Zhang, Q.; Liu, H.; Nie, L. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell 2020, 182, 1284–1294.e1289.
- Andreano, E.; Piccini, G.; Licastro, D.; Casalino, L.; Johnson, N.V.; Paciello, I.; Dal Monego, S.; Pantano, E.; Manganaro, N.; Manenti, A. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv 2020.
- Cai, Y.; Zhang, J.; Xiao, T.; Lavine, C.L.; Rawson, S.; Peng, H.; Zhu, H.; Anand, K.; Tong, P.; Gautam, A. Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants. bioRxiv 2021.
- Liu, Z.; VanBlargan, L.A.; Rothlauf, P.W.; Bloyet, L.-M.; Chen, R.E.; Stumpf, S.; Zhao, H.; Errico, J.M.; Theel, E.S.; Ellebedy, A. Landscape analysis of escape variants identifies SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. SSRN Electron. J. 2020.
- Greaney, A.J.; Starr, T.N.; Gilchuk, P.; Zost, S.J.; Binshtein, E.; Loes, A.N.; Hilton, S.K.; Huddleston, J.; Eguia, R.; Crawford, K.H. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe 2021, 29, 44–57.e49.
- Starr, T.N.; Greaney, A.J.; Dingens, A.S.; Bloom, J.D. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep. Med. 2021, 2, 100255.
- Starr, T.N.; Greaney, A.J.; Addetia, A.; Hannon, W.W.; Choudhary, M.C.; Dingens, A.S.; Li, J.Z.; Bloom, J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science 2021, 371, 850–854.
- Barnes, C.O.; Jette, C.A.; Abernathy, M.E.; Dam, K.-M.A.; Esswein, S.R.; Gristick, H.B.; Malyutin, A.G.; Sharaf, N.G.; Huey-Tubman, K.E.; Lee, Y.E. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 2020, 588, 682–687.
- Piccoli, L.; Park, Y.-J.; Tortorici, M.A.; Czudnochowski, N.; Walls, A.C.; Beltramello, M.; Silacci-Fregni, C.; Pinto, D.; Rosen, L.E.; Bowen, J.E. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell 2020, 183, 1024–1042.e1021.
- Baum, A.; Fulton, B.O.; Wloga, E.; Copin, R.; Pascal, K.E.; Russo, V.; Giordano, S.; Lanza, K.; Negron, N.; Ni, M. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 2020, 369, 1014–1018.
- Rees-Spear, C.; Muir, L.; Griffith, S.A.; Heaney, J.; Aldon, Y.; Snitselaar, J.L.; Thomas, P.; Graham, C.; Seow, J.; Lee, N.; et al. The effect of spike mutations on SARS-CoV-2 neutralization. Cell Rep. 2021, 34, 108890.
- Tada, T.; Dcosta, B.M.; Samanovic-Golden, M.; Herati, R.S.; Cornelius, A.; Mulligan, M.J.; Landau, N.R. Neutralization of viruses with European, South African, and United States SARS-CoV-2 variant spike proteins by convalescent sera and BNT162b2 mRNA vaccine-elicited antibodies. bioRxiv 2021.
- Wu, K.; Werner, A.P.; Koch, M.; Choi, A.; Narayanan, E.; Stewart-Jones, G.B.; Colpitts, T.; Bennett, H.; Boyoglu-Barnum, S.; Shi, W. Serum neutralizing activity elicited by mRNA-1273 vaccine. N. Engl. J. Med. 2021, 384, 1468–1470.
- Robbiani, D.F.; Gaebler, C.; Muecksch, F.; Lorenzi, J.C.; Wang, Z.; Cho, A.; Agudelo, M.; Barnes, C.O.; Gazumyan, A.; Finkin, S. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020, 584, 437–442.
- Yuan, M.; Liu, H.; Wu, N.C.; Lee, C.-C.D.; Zhu, X.; Zhao, F.; Huang, D.; Yu, W.; Hua, Y.; Tien, H. Structural basis of a shared antibody response to SARS-CoV-2. Science 2020, 369, 1119–1123.
- Zost, S.J.; Gilchuk, P.; Chen, R.E.; Case, J.B.; Reidy, J.X.; Trivette, A.; Nargi, R.S.; Sutton, R.E.; Suryadevara, N.; Chen, E.C. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020, 26, 1422–1427.
- Steffen, T.L.; Stone, E.T.; Hassert, M.; Geerling, E.; Grimberg, B.T.; Espino, A.M.; Pantoja, P.; Climent, C.; Hoft, D.F.; George, S.L.; et al. The receptor binding domain of SARS-CoV-2 spike is the key target of neutralizing antibody in human polyclonal sera. bioRxiv 2020.
- Rodda, L.B.; Netland, J.; Shehata, L.; Pruner, K.B.; Morawski, P.A.; Thouvenel, C.D.; Takehara, K.K.; Eggenberger, J.; Hemann, E.A.; Waterman, H.R. Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell 2021, 184, 169–183.e117.
- Wajnberg, A.; Amanat, F.; Firpo, A.; Altman, D.R.; Bailey, M.J.; Mansour, M.; McMahon, M.; Meade, P.; Mendu, D.R.; Muellers, K. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science 2020, 370, 1227–1230.
- Liu, Z.; VanBlargan, L.A.; Bloyet, L.-M.; Rothlauf, P.W.; Chen, R.E.; Stumpf, S.; Zhao, H.; Errico, J.M.; Theel, E.S.; Liebeskind, M.J. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 2021, 29, 477–488.e474.
- Collier, D.A.; De Marco, A.; Ferreira, I.A.; Meng, B.; Datir, R.; Walls, A.C.; Bassi, J.; Pinto, D.; Fregni, C.S.; Bianchi, S. SARS-CoV-2 B. 1.1. 7 sensitivity to mRNA vaccine-elicited, convalescent and monoclonal antibodies. medRxiv 2021.
- Muik, A.; Wallisch, A.-K.; Sänger, B.; Swanson, K.A.; Mühl, J.; Chen, W.; Cai, H.; Maurus, D.; Sarkar, R.; Türeci, Ö. Neutralization of SARS-CoV-2 lineage B. 1.1. 7 pseudovirus by BNT162b2 vaccine–elicited human sera. Science 2021, 371, 1152–1153.
- Yadav, P.D.; Sapkal, G.; Ella, R.; Sahay, R.R.; Nyayanit, D.A.; Patil, D.Y.; Deshpande, G.; Shete, A.M.; Gupta, N.; Mohan, V.K. Neutralization against B. 1.351 and B. 1.617. 2 with sera of COVID-19 recovered cases and vaccinees of BBV152. bioRxiv 2021.
- Anderson, E.J.; Rouphael, N.G.; Widge, A.T.; Jackson, L.A.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. 2020, 383, 2427–2438.
- Liu, L.; Wang, P.; Nair, M.S.; Yu, J.; Rapp, M.; Wang, Q.; Luo, Y.; Chan, J.F.-W.; Sahi, V.; Figueroa, A. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 2020, 584, 450–456.
- Baay, M.; Lina, B.; Fontanet, A.; Marchant, A.; Saville, M.; Sabot, P.; Duclos, P.; Vandeputte, J.; Neels, P. SARS-CoV-2: Virology, epidemiology, immunology and vaccine development. Biologicals 2020, 66, 35–40.
- Logunov, D.Y.; Dolzhikova, I.V.; Zubkova, O.V.; Tukhvatullin, A.I.; Shcheblyakov, D.V.; Dzharullaeva, A.S.; Grousova, D.M.; Erokhova, A.S.; Kovyrshina, A.V.; Botikov, A.G. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: Two open, non-randomised phase 1/2 studies from Russia. Lancet 2020, 396, 887–897.
- Salvatori, G.; Luberto, L.; Maffei, M.; Aurisicchio, L.; Roscilli, G.; Palombo, F.; Marra, E. SARS-CoV-2 SPIKE PROTEIN: An optimal immunological target for vaccines. J. Transl. Med. 2020, 18, 1–3.
- WHO Issues Its First Emergency Use Validation for a COVID-19 Vaccine and Emphasizes Need for Equitable Global Access. Available online: https://www.who.int/news/item/31-12-2020-who-issues-its-first-emergency-use-validation-for-a-covid-19-vaccine-and-emphasizes-need-for-equitable-global-access (accessed on 16 June 2021).
- Rubin, R. COVID-19 Vaccines vs. Variants—Determining How Much Immunity Is Enough. JAMA 2021, 325, 1241–1243.
- Moore, J.P.; Offit, P.A. SARS-CoV-2 vaccines and the growing threat of viral variants. JAMA 2021, 325, 821–822.
- Weissman, D.; Alameh, M.-G.; de Silva, T.; Collini, P.; Hornsby, H.; Brown, R.; LaBranche, C.C.; Edwards, R.J.; Sutherland, L.; Santra, S. D614G spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host Microbe 2021, 29, 23–31.e24.
- Moderna Announces It Has Shipped Variant-Specific Vaccine Candidate, mRNA-1273.351, to NIH for Clinical Study. Available online: https://investors.modernatx.com/news-releases/news-release-details/moderna-announces-it-has-shipped-variant-specific-vaccine (accessed on 16 June 2021).
- Novavax COVID-19 Vaccine Demonstrates 89.3% Efficacy in UK Phase 3 Trial. Available online: https://ir.novavax.com/news-releases/news-release-details/novavax-covid-19-vaccine-demonstrates-893-efficacy-uk-phase-3 (accessed on 20 June 2021).
- Pfizer and BioNTech Initiate a Study as Part of Broad Development Plan to Evaluate COVID-19 Booster and New Vaccine Variants. Available online: https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-initiate-study-part-broad-development (accessed on 20 June 2021).
- Mahase, E. Covid-19: Novavax vaccine efficacy is 86% against UK variant and 60% against South African variant. BMJ 2021.
- Wise, J. Covid-19: The E484K mutation and the risks it poses. BMJ 2021.
- Abu-Raddad, L.J.; Chemaitelly, H.; Butt, A.A. Effectiveness of the BNT162b2 Covid-19 Vaccine against the B. 1.1. 7 and B. 1.351 Variants. N. Engl. J. Med. 2021, 385, 187–189.
- Thiagarajan, K. Why is India having a covid-19 surge? BMJ 2021.
- de Souza, W.M.; Amorim, M.R.; Sesti-Costa, R.; Coimbra, L.D.; Toledo-Teixeira, D.A.d.; Parise, P.L.; Barbosa, P.P.; Bispo-dos-Santos, K.; Mofatto, L.S.; Simeoni, C.L.; et al. Levels of SARS-CoV-2 lineage P. 1 neutralization by antibodies elicited after natural infection and vaccination. SSRN Electron. J. 2021. Available online: https://ssrn.com/abstract=3793486 (accessed on 20 June 2021).
- Bandoy, D.D.R.; Weimer, B.C. Analysis of SARS-CoV-2 genomic epidemiology reveals disease transmission coupled to variant emergence and allelic variation. Sci. Rep. 2021, 11, 1–12.
- Hoo, H.E.; Loh, H.C.; Ch’ng, A.S.H.; Hoo, F.K.; Looi, I. Positive impacts of the COVID-19 pandemic and public health measures on healthcare. Prog. Microbes Mol. Biol. 2021, 4.




