
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Annika Karger | + 2975 word(s) | 2975 | 2021-08-24 11:18:18 | | | |
| 2 | Vivi Li | Meta information modification | 2975 | 2021-09-29 11:57:03 | | |
Video Upload Options
Ever since RNA sequencing of whole genomes and transcriptomes became available, numerous RNA transcripts without having the classic function of encoding proteins have been discovered. Long non-coding RNAs (lncRNAs) with a length greater than 200 nucleotides were considered as “junk” in the beginning, but it has increasingly become clear that lncRNAs have crucial roles in regulating a variety of cellular mechanisms and are often deregulated in several diseases, such as cancer. Lung cancer is the leading cause of cancer-related deaths and has a survival rate of less than 10%. Immune cells infiltrating the tumor microenvironment (TME) have been shown to have a great effect on tumor development with macrophages being the major cell type within the TME. Macrophages can inherit an inflammatory M1 or an anti-inflammatory M2 phenotype. Tumor-associated macrophages, which are predominantly polarized to M2, favor tumor growth, angiogenesis, and metastasis.
1. Lung Cancer
2. Long Non-Coding RNAs
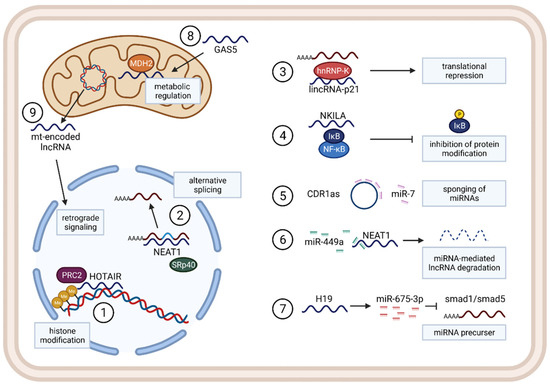
2.1. Functions and Mechanisms
2.2. LncRNAs in Cancer
2.3. LncRNAs in Immunity, Inflammation, and the TME
2.4. LncRNAs in Macrophages in Lung Cancer
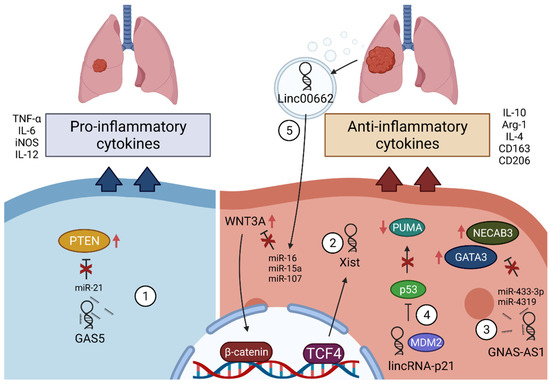
2.4.1. GAS5
2.4.2. Xist
2.4.3. GNAS-AS1
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30.
- Travis, W.D.; Brambilla, E.; Riely, G.J. New pathologic classification of lung cancer: Relevance for clinical practice and clinical trials. J. Clin. Oncol. 2013, 31, 992–1001.
- Travis, W.D.; Brambilla, E.; Nicholson, A.G.; Yatabe, Y.; Austin, J.H.M.; Beasley, M.B.; Chirieac, L.R.; Dacic, S.; Duhig, E.; Flieder, D.B.; et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J. Thorac. Oncol. 2015, 10, 1243–1260.
- Inamura, K. Lung Cancer: Understanding Its Molecular Pathology and the 2015 WHO Classification. Front. Oncol. 2017, 7, 193.
- Baxevanos, P.; Mountzios, G. Novel chemotherapy regimens for advanced lung cancer: Have we reached a plateau? Ann. Transl. Med. 2018, 6, 139.
- Seebacher, N.A.; Stacy, A.E.; Porter, G.M.; Merlot, A.M. Clinical development of targeted and immune based anti-cancer therapies. J. Exp. Clin. Cancer Res. 2019, 38, 156.
- Whiteside, T.L. Immune suppression in cancer: Effects on immune cells, mechanisms and future therapeutic intervention. Semin. Cancer Biol. 2006, 16, 3–15.
- Sumitomo, R.; Hirai, T.; Fujita, M.; Murakami, H.; Otake, Y.; Huang, C.L. PD-L1 expression on tumor-infiltrating immune cells is highly associated with M2 TAM and aggressive malignant potential in patients with resected non-small cell lung cancer. Lung Cancer 2019, 136, 136–144.
- Chen, D.S.; Mellman, I. Oncology meets immunology: The cancer-immunity cycle. Immunity 2013, 39, 1–10.
- Groth, C.; Hu, X.; Weber, R.; Fleming, V.; Altevogt, P.; Utikal, J.; Umansky, V. Immunosuppression mediated by myeloid-derived suppressor cells (MDSCs) during tumour progression. Br. J. Cancer 2019, 120, 16–25.
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012, 366, 2443–2454.
- Ock, C.Y.; Hwang, J.E.; Keam, B.; Kim, S.B.; Shim, J.J.; Jang, H.J.; Park, S.; Sohn, B.H.; Cha, M.; Ajani, J.A.; et al. Genomic landscape associated with potential response to anti-CTLA-4 treatment in cancers. Nat. Commun. 2017, 8, 1050.
- Robert, C. A decade of immune-checkpoint inhibitors in cancer therapy. Nat. Commun. 2020, 11, 3801.
- Jin, J.; Wu, X.; Yin, J.; Li, M.; Shen, J.; Li, J.; Zhao, Y.; Zhao, Q.; Wu, J.; Wen, Q.; et al. Identification of Genetic Mutations in Cancer: Challenge and Opportunity in the New Era of Targeted Therapy. Front. Oncol. 2019, 9, 263.
- Normanno, N.; Barberis, M.; De Marinis, F.; Gridelli, C.; On the Behalf of the Aiot Expert Panel. Molecular and Genomic Profiling of Lung Cancer in the Era of Precision Medicine: A Position Paper from the Italian Association of Thoracic Oncology (AIOT). Cancers 2020, 12, 1627.
- Sholl, L.M.; Aisner, D.L.; Varella-Garcia, M.; Berry, L.D.; Dias-Santagata, D.; Wistuba, I.I.; Chen, H.; Fujimoto, J.; Kugler, K.; Franklin, W.A.; et al. Multi-institutional Oncogenic Driver Mutation Analysis in Lung Adenocarcinoma: The Lung Cancer Mutation Consortium Experience. J. Thorac. Oncol. 2015, 10, 768–777.
- Nan, X.; Xie, C.; Yu, X.; Liu, J. EGFR TKI as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer. Oncotarget 2017, 8, 75712–75726.
- Yuan, M.; Huang, L.L.; Chen, J.H.; Wu, J.; Xu, Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct. Target. Ther. 2019, 4, 61.
- Ghafouri-Fard, S.; Shoorei, H.; Branicki, W.; Taheri, M. Non-coding RNA profile in lung cancer. Exp. Mol. Pathol. 2020, 114, 104411.
- Lai, X.N.; Li, J.; Tang, L.B.; Chen, W.T.; Zhang, L.; Xiong, L.X. MiRNAs and LncRNAs: Dual Roles in TGF-beta Signaling-Regulated Metastasis in Lung Cancer. Int. J. Mol. Sci. 2020, 21, 1193.
- Peng, F.; Wang, R.; Zhang, Y.; Zhao, Z.; Zhou, W.; Chang, Z.; Liang, H.; Zhao, W.; Qi, L.; Guo, Z.; et al. Differential expression analysis at the individual level reveals a lncRNA prognostic signature for lung adenocarcinoma. Mol. Cancer 2017, 16, 98.
- Gibb, E.A.; Warren, R.L.; Wilson, G.W.; Brown, S.D.; Robertson, G.A.; Morin, G.B.; Holt, R.A. Activation of an endogenous retrovirus-associated long non-coding RNA in human adenocarcinoma. Genome Med. 2015, 7, 22.
- Tian, X.; Wu, Y.; Yang, Y.; Wang, J.; Niu, M.; Gao, S.; Qin, T.; Bao, D. Long noncoding RNA LINC00662 promotes M2 macrophage polarization and hepatocellular carcinoma progression via activating Wnt/β-catenin signaling. Mol. Oncol. 2020, 14, 462–483.
- Liu, J.; Ding, D.; Jiang, Z.; Du, T.; Liu, J.; Kong, Z. Long non-coding RNA CCAT1/miR-148a/PKCζ prevents cell migration of prostate cancer by altering macrophage polarization. Prostate 2019, 79, 105–112.
- Cao, J.; Dong, R.; Jiang, L.; Gong, Y.; Yuan, M.; You, J.; Meng, W.; Chen, Z.; Zhang, N.; Weng, Q.; et al. LncRNA-MM2P Identified as a Modulator of Macrophage M2 Polarization. Cancer Immunol. Res. 2019, 7, 292–305.
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108.
- Santosh, B.; Varshney, A.; Yadava, P.K. Non-coding RNAs: Biological functions and applications. Cell Biochem. Funct. 2015, 33, 14–22.
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693.
- Cooper, D.R.; Carter, G.; Li, P.; Patel, R.; Watson, J.E.; Patel, N.A. Long Non-Coding RNA NEAT1 Associates with SRp40 to Temporally Regulate PPARgamma2 Splicing during Adipogenesis in 3T3-L1 Cells. Genes 2014, 5, 1050–1063.
- Rashid, F.; Shah, A.; Shan, G. Long Non-coding RNAs in the Cytoplasm. Genom. Proteom. Bioinform. 2016, 14, 73–80.
- Liu, B.; Sun, L.; Liu, Q.; Gong, C.; Yao, Y.; Lv, X.; Lin, L.; Yao, H.; Su, F.; Li, D.; et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 2015, 27, 370–381.
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388.
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338.
- Dey, B.K.; Pfeifer, K.; Dutta, A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014, 28, 491–501.
- Sang, L.; Ju, H.Q.; Yang, Z.; Ge, Q.; Zhang, Z.; Liu, F.; Yang, L.; Gong, H.; Shi, C.; Qu, L.; et al. Mitochondrial long non-coding RNA GAS5 tunes TCA metabolism in response to nutrient stress. Nat. Metab. 2021, 3, 90–106.
- Zhao, Y.; Sun, L.; Wang, R.R.; Hu, J.F.; Cui, J. The effects of mitochondria-associated long noncoding RNAs in cancer mitochondria: New players in an old arena. Crit. Rev. Oncol. Hematol. 2018, 131, 76–82.
- Zhou, Y.; Zhong, Y.; Wang, Y.; Zhang, X.; Batista, D.L.; Gejman, R.; Ansell, P.J.; Zhao, J.; Weng, C.; Klibanski, A. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 2007, 282, 24731–24742.
- Betts, J.A.; Moradi Marjaneh, M.; Al-Ejeh, F.; Lim, Y.C.; Shi, W.; Sivakumaran, H.; Tropee, R.; Patch, A.M.; Clark, M.B.; Bartonicek, N.; et al. Long Noncoding RNAs CUPID1 and CUPID2 Mediate Breast Cancer Risk at 11q13 by Modulating the Response to DNA Damage. Am. J. Hum. Genet. 2017, 101, 255–266.
- Liu, X.; Xiao, Z.D.; Han, L.; Zhang, J.; Lee, S.W.; Wang, W.; Lee, H.; Zhuang, L.; Chen, J.; Lin, H.K.; et al. LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat. Cell Biol. 2016, 18, 431–442.
- Li, S.; Mei, Z.; Hu, H.B.; Zhang, X. The lncRNA MALAT1 contributes to non-small cell lung cancer development via modulating miR-124/STAT3 axis. J. Cell. Physiol. 2018, 233, 6679–6688.
- Zhou, X.; Liu, S.; Cai, G.; Kong, L.; Zhang, T.; Ren, Y.; Wu, Y.; Mei, M.; Zhang, L.; Wang, X. Long Non Coding RNA MALAT1 Promotes Tumor Growth and Metastasis by inducing Epithelial-Mesenchymal Transition in Oral Squamous Cell Carcinoma. Sci. Rep. 2015, 5, 15972.
- Ji, P.; Diederichs, S.; Wang, W.; Boing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041.
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076.
- Padua Alves, C.; Fonseca, A.S.; Muys, B.R.; de Barros, E.L.B.R.; Burger, M.C.; de Souza, J.E.; Valente, V.; Zago, M.A.; Silva, W.A., Jr. Brief report: The lincRNA Hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cells 2013, 31, 2827–2832.
- Kim, K.; Jutooru, I.; Chadalapaka, G.; Johnson, G.; Frank, J.; Burghardt, R.; Kim, S.; Safe, S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 2013, 32, 1616–1625.
- Ren, Y.; Jia, H.H.; Xu, Y.Q.; Zhou, X.; Zhao, X.H.; Wang, Y.F.; Song, X.; Zhu, Z.Y.; Sun, T.; Dou, Y.; et al. Paracrine and epigenetic control of CAF-induced metastasis: The role of HOTAIR stimulated by TGF-ss1 secretion. Mol. Cancer 2018, 17, 5.
- Chen, Z.Z.; Huang, L.; Wu, Y.H.; Zhai, W.J.; Zhu, P.P.; Gao, Y.F. LncSox4 promotes the self-renewal of liver tumour-initiating cells through Stat3-mediated Sox4 expression. Nat. Commun. 2016, 7, 12598.
- Zhu, P.; Wang, Y.; Wu, J.; Huang, G.; Liu, B.; Ye, B.; Du, Y.; Gao, G.; Tian, Y.; He, L.; et al. LncBRM initiates YAP1 signalling activation to drive self-renewal of liver cancer stem cells. Nat. Commun. 2016, 7, 13608.
- Mowel, W.K.; Kotzin, J.J.; McCright, S.J.; Neal, V.D.; Henao-Mejia, J. Control of Immune Cell Homeostasis and Function by lncRNAs. Trends Immunol. 2018, 39, 55–69.
- Chen, J.; Ao, L.; Yang, J. Long non-coding RNAs in diseases related to inflammation and immunity. Ann. Transl. Med. 2019, 7, 494.
- Hu, G.; Tang, Q.; Sharma, S.; Yu, F.; Escobar, T.M.; Muljo, S.A.; Zhu, J.; Zhao, K. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat. Immunol. 2013, 14, 1190–1198.
- Pang, K.C.; Dinger, M.E.; Mercer, T.R.; Malquori, L.; Grimmond, S.M.; Chen, W.; Mattick, J.S. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J. Immunol. 2009, 182, 7738–7748.
- Xia, F.; Dong, F.; Yang, Y.; Huang, A.; Chen, S.; Sun, D.; Xiong, S.; Zhang, J. Dynamic transcription of long non-coding RNA genes during CD4+ T cell development and activation. PLoS ONE 2014, 9, e101588.
- Rapicavoli, N.A.; Qu, K.; Zhang, J.; Mikhail, M.; Laberge, R.M.; Chang, H.Y. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. eLife 2013, 2, e00762.
- Zhang, Y.; Feng, J.; Fu, H.; Liu, C.; Yu, Z.; Sun, Y.; She, X.; Li, P.; Zhao, C.; Liu, Y.; et al. Coagulation Factor X Regulated by CASC2c Recruited Macrophages and Induced M2 Polarization in Glioblastoma Multiforme. Front. Immunol. 2018, 9, 1557.
- Jiang, R.; Tang, J.; Chen, Y.; Deng, L.; Ji, J.; Xie, Y.; Wang, K.; Jia, W.; Chu, W.M.; Sun, B. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat. Commun. 2017, 8, 15129.
- Sharma, N.S.; Gnamlin, P.; Durden, B.; Gupta, V.K.; Kesh, K.; Garrido, V.T.; Dudeja, V.; Saluja, A.; Banerjee, S. Long non-coding RNA GAS5 acts as proliferation “brakes” in CD133+ cells responsible for tumor recurrence. Oncogenesis 2019, 8, 68.
- Wang, Y.; Xin, S.; Zhang, K.; Shi, R.; Bao, X. Low GAS5 Levels as a Predictor of Poor Survival in Patients with Lower-Grade Gliomas. J. Oncol. 2019, 2019, 1785042.
- Shen, J.; Hodges, T.R.; Song, R.; Gong, Y.; Calin, G.A.; Heimberger, A.B.; Zhao, H. Serum HOTAIR and GAS5 levels as predictors of survival in patients with glioblastoma. Mol. Carcinog. 2018, 57, 137–141.
- Sun, M.; Jin, F.; Xia, R.; Kong, R.; Li, J.; Xu, T.; Liu, Y.; Zhang, E.; Liu, X.; De, W. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer 2014, 319, 1–12.
- Lambrou, G.I.; Hatziagapiou, K.; Zaravinos, A. The Non-Coding RNA GAS5 and Its Role in Tumor Therapy-Induced Resistance. Int. J. Mol. Sci. 2020, 21, 7633.
- Shi, X.; Sun, M.; Liu, H.; Yao, Y.; Kong, R.; Chen, F.; Song, Y. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol. Carcinog. 2015, 54 (Suppl. S1), E1–E12.
- Cao, L.; Chen, J.; Ou, B.; Liu, C.; Zou, Y.; Chen, Q. GAS5 knockdown reduces the chemo-sensitivity of non-small cell lung cancer (NSCLC) cell to cisplatin (DDP) through regulating miR-21/PTEN axis. Biomed. Pharmacother. 2017, 93, 570–579.
- Wang, X.; Li, F.Y.; Zhao, W.; Gao, Z.K.; Shen, B.; Xu, H.; Cui, Y.F. Long non-coding RNA GAS5 overexpression inhibits M2-like polarization of tumour-associated macrophages in SMCC-7721 cells by promoting PTEN expression. Int. J. Exp. Pathol. 2020, 101, 215–222.
- Zhang, Y.; Lu, X.; Yang, M.; Shangguan, J.; Yin, Y. GAS5 knockdown suppresses inflammation and oxidative stress induced by oxidized low-density lipoprotein in macrophages by sponging miR-135a. Mol. Cell Biochem. 2021, 476, 949–957.
- Sahin, E.; Haubenwallner, S.; Kuttke, M.; Kollmann, I.; Halfmann, A.; Dohnal, A.M.; Chen, L.; Cheng, P.; Hoesel, B.; Einwallner, E.; et al. Macrophage PTEN regulates expression and secretion of arginase I modulating innate and adaptive immune responses. J. Immunol. 2014, 193, 1717–1727.
- Li, N.; Qin, J.; Lan, L.; Zhang, H.; Liu, F.; Wu, Z.; Ni, H.; Wang, Y. PTEN inhibits macrophage polarization from M1 to M2 through CCL2 and VEGF-A reduction and NHERF-1 synergism. Cancer Biol. Ther. 2015, 16, 297–306.
- Sarode, P.; Zheng, X.; Giotopoulou, G.A.; Weigert, A.; Kuenne, C.; Günther, S.; Friedrich, A.; Gattenlöhner, S.; Stiewe, T.; Brüne, B.; et al. Reprogramming of tumor-associated macrophages by targeting β-catenin/FOSL2/ARID5A signaling: A potential treatment of lung cancer. Sci. Adv. 2020, 6, eaaz6105.
- Sahakyan, A.; Yang, Y.; Plath, K. The Role of Xist in X-Chromosome Dosage Compensation. Trends Cell Biol. 2018, 28, 999–1013.
- Sun, W.; Zu, Y.; Fu, X.; Deng, Y. Knockdown of lncRNA-XIST enhances the chemosensitivity of NSCLC cells via suppression of autophagy. Oncol Rep. 2017, 38, 3347–3354.
- Tian, L.J.; Wu, Y.P.; Wang, D.; Zhou, Z.H.; Xue, S.B.; Zhang, D.Y.; Wei, Y.G.; Liu, W. Upregulation of Long Noncoding RNA (lncRNA) X-Inactive Specific Transcript (XIST) is Associated with Cisplatin Resistance in Non-Small Cell Lung Cancer (NSCLC) by Downregulating MicroRNA-144-3p. Med. Sci. Monit. 2019, 25, 8095–8104.
- Li, L.; Lv, G.; Wang, B.; Kuang, L. XIST/miR-376c-5p/OPN axis modulates the influence of proinflammatory M1 macrophages on osteoarthritis chondrocyte apoptosis. J. Cell Physiol. 2020, 235, 281–293.
- Zhao, Y.; Yu, Z.; Ma, R.; Zhang, Y.; Zhao, L.; Yan, Y.; Lv, X.; Zhang, L.; Su, P.; Bi, J.; et al. lncRNA-Xist/miR-101-3p/KLF6/C/EBPalpha axis promotes TAM polarization to regulate cancer cell proliferation and migration. Mol. Ther. Nucleic Acids 2021, 23, 536–551.
- Sun, Y.; Xu, J. TCF-4 Regulated lncRNA-XIST Promotes M2 Polarization Of Macrophages And Is Associated With Lung Cancer. Onco Targets Ther. 2019, 12, 8055–8062.
- Wang, X.W.; Xu, H.; Wang, C.H.; Xie, H. Long non-coding RNA GNAS-AS1 promotes cell migration and invasion via regulating Wnt/β-catenin pathway in nasopharyngeal carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 3077–3084.
- Li, Z.; Feng, C.; Guo, J.; Hu, X.; Xie, D. GNAS-AS1/miR-4319/NECAB3 axis promotes migration and invasion of non-small cell lung cancer cells by altering macrophage polarization. Funct. Integr. Genom. 2020, 20, 17–28.
- Liu, S.Q.; Zhou, Z.Y.; Dong, X.; Guo, L.; Zhang, K.J. LncRNA GNAS-AS1 facilitates ER+ breast cancer cells progression by promoting M2 macrophage polarization via regulating miR-433-3p/GATA3 axis. Biosci. Rep. 2020, 40, BSR20200626.
- Nakaoka, H.J.; Hara, T.; Yoshino, S.; Kanamori, A.; Matsui, Y.; Shimamura, T.; Sato, H.; Murakami, Y.; Seiki, M.; Sakamoto, T. NECAB3 Promotes Activation of Hypoxia-inducible factor-1 during Normoxia and Enhances Tumourigenicity of Cancer Cells. Sci. Rep. 2016, 6, 22784.




