Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Thierry Naas | + 1202 word(s) | 1202 | 2021-09-13 08:27:48 | | | |
| 2 | Amina Yu | Meta information modification | 1202 | 2021-09-23 11:02:06 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Naas, T. Antimicrobial Peptides and Antibiotics. Encyclopedia. Available online: https://encyclopedia.pub/entry/14454 (accessed on 07 February 2026).
Naas T. Antimicrobial Peptides and Antibiotics. Encyclopedia. Available at: https://encyclopedia.pub/entry/14454. Accessed February 07, 2026.
Naas, Thierry. "Antimicrobial Peptides and Antibiotics" Encyclopedia, https://encyclopedia.pub/entry/14454 (accessed February 07, 2026).
Naas, T. (2021, September 23). Antimicrobial Peptides and Antibiotics. In Encyclopedia. https://encyclopedia.pub/entry/14454
Naas, Thierry. "Antimicrobial Peptides and Antibiotics." Encyclopedia. Web. 23 September, 2021.
Copy Citation
Antimicrobial peptides constitute one of the most promising alternatives to antibiotics since they could be used to treat bacterial infections, especially those caused by multidrug-resistant pathogens. Many antimicrobial peptides, with various activity spectra and mechanisms of actions, have been described.
antimicrobial peptides
antibiotic resistance
multidrug resistance
ESKAPE
1. Introduction
There was collective enthusiasm about the advent of antibiotic therapy, and this optimism prevailed in the light of the constant discoveries of new classes of antibiotics, despite the very early description of therapeutic failures and resistance to treatments with these drugs [1]. At the present, in 2021, multidrug-resistant (MDR) and even pan-drug-resistant (PDR) bacteria have spread widely around the world and are currently responsible for increasing morbidity and mortality rates, as well as the significant cost to society [2]. The low frequency of discoveries of new classes of antibiotics, and the rapid emergence of resistance to novel antibiotics, show the need for novel therapeutic alternatives to antibiotics, such as lysine-based small molecules, vaccines, anti-virulence strategies, phage therapy and antimicrobial peptides.
Antimicrobial resistance (AMR) is an emerging global health problem that results, in some cases, in difficulties to treat bacterial infections. It was listed by the World Health Organization (WHO) among the top ten global public health threats facing humanity, as it is predicted to cause about 10 million deaths each year by 2050 [3]. Therefore, efforts to slow down the propagation of AMR have been implemented worldwide. As such, a Global Action Plan on Antimicrobial Resistance (GAP) was created in 2015, aiming to implement national action plans to limit the progress of AMR. In addition, the WHO reports call for urgent action to avert an antimicrobial resistance crisis and insists on the importance of discovering and developing new antibiotics. Thus, new compounds that are active against pathogens, especially those which cause nosocomial infections and tend to adopt multidrug resistance, are needed. To curb this problem, several alternative therapies have been proposed, among which antimicrobial peptides (AMPs) were suggested to be very promising more than 20 years ago, as they have existed in nature for millions of years with almost no or limited resistance development [4]. This makes them very attractive compared to antibiotics that develop resistance relatively fast. This absence/slow development of resistance against microbes may be attributed to the presence of various modes of action of AMPs against the bacteria in comparison to the fixed targets used by the antibiotics [5]. In addition, AMPs are considered less toxic, as they are broken down into amino acids unlike other therapeutics, which might generate potentially harmful metabolites. This review is to highlight where these molecules stand now in the overall scheme to curb MDR bacterial infections. Their potential to counteract AMR, to replace traditional antibiotics, to evaluate their benefits and to describe the challenges faced by R&D will be discussed in this review.
AMPs are small polypeptide molecules, typically made up of around 12 to 50 amino acids, found in all classes of living organisms [6]. These molecules are produced as secondary metabolites, are part of the innate immunity and are, in mammals, usually ribosomally produced by epithelial cells, but also by phagocytes (cells of the immune system). These peptides can be found in tissues or mucous membranes; in fact, the latter harbor a multitude of pathogenic or commensal microorganisms. Among these peptides, some have a broad spectrum of antimicrobial activity, capable of inhibiting or killing different types of microorganisms (MO) (Gram-positive or -negative bacteria and/or fungi) but also protozoans, and viruses [7][8]. According to The Antimicrobial Peptide Database, more than 3257 antimicrobial peptides have been described to date [9]. They originate from six kingdoms, where 365 come from bacteria, 5 from archaea, 8 from protists, 22 from fungi, 360 from plants, and 2414 from animals, in addition to some synthetic peptides. Recently, another webserver with several subsections that screen among 40,000 entries has been made available. It summarizes all available data for desired peptide properties, thus allowing further structural and functional studies [10].
2. Classification and Mode of Action of Antimicrobial Peptides
AMPs are usually classified according to several criteria: First, based on their biological source. In this classification, we distinguish AMPs from human and mammalian sources, such as cathelicidin and defensin, which make up the most important families of AMPs [11]; AMPs from amphibians [12], fish [13], insects [14], and plants [15] are also classified in this category. Classification could be based on the AMP’s biological functions; for example, antibacterial, antiviral, antifungal, antiparasitic peptides were described. Third, based on their biochemical properties (amino acid sequence, composition, length, hydrophobicity, charge), where conformation and structure serve as criteria for classification as well [7].
3. AMPs for Diagnostic Purposes
Rapid diagnostic tools are the key to prevent the spread of contagious infectious agents by rapidly identifying carriers or infected patients and prompt implementation of adapted infection control measures. The ability of AMPs to sense bacterial presence in potentially infected clinical samples is a major advantage. As already reviewed by Pardoux et al., AMPs could serve as infection selective tracers where they act as probes in biosensors and thus detect pathogens [16]. This is due to the preferential binding of AMPs along with their great number allowing the coverage of a wide range of microorganisms. AMPs were also described as powerful radiotracers and proved to be even better than labeled antibiotics [17]. [99mTc-HYNIC/EDDA]-MccJ25 is an example of an antimicrobial peptide analog that was used as a potential radiotracer for detection of infection [16]. Interestingly, this imaging agent for E. coli infection is also stable against enzymatic and metabolic degradation [18].
4. AMPs Facing 2019 Pandemic: SARS-CoV-2
In addition to activity toward bacteria, according to the database of antimicrobial activity and structure of peptides (DBAASP) [19]., 208 AMPs have been reported to show antiviral activities against SARS-CoV-2. Slight changes to glycocin F and lactococcin G increased their efficacy against SARS-CoV-2 by inhibiting protein synthesis with no side effects [20]. Similarly, mucroporin-M1 (LFRLIKSLIKRLVSAFK), a peptide analog displaying four amino acid changes (G3R, P6K, G10K, and G11R) as compared to the parent peptide mucroporin shows therapeutic activities against SARS-CoV-2 [21]. Brilacidin and LF are additional drug candidates for SARS-CoV-2 treatment [22][23]. Caerin 1.6 and caerin 1.10, two other AMPs, have a very high potential to interact with the spike surface protein of SARS-CoV-2, but low affinity for the ACE2 receptor. The selectivity of these peptides toward viral proteins makes them potential candidates for SARS-CoV-2 therapy [24].
5. Conclusions
AMPs constitute promising agents with good antibacterial activity against a large set of pathogens, including Gram-negative and Gram-positive bacteria. Their ability to kill ESKAPE pathogens, especially those being pan-drug-resistant, represents a clinical added value over antibiotics due to the limited treatment options available to eradicate such infections. In this review, examples of AMPs effective against MDR bacteria that have the potential to replace conventional antibiotics have been reported. Their roles in boosting the activity of therapeutic molecules, eradication of biofilms, and preventive healthcare broaden AMPs’ therapeutic potential (Figure 1). However, despite the huge number of AMPs described, and the efforts to optimize them, only a few reached advanced clinical trials. There are several challenges, such as their low stability, high extraction costs, and cytotoxicity, which need to be overcome so AMPs can be used in clinics. Nevertheless, due to increasing research efforts, it is expected that, hopefully, potent alternatives to antibacterial agents with the ability to eradicate untreatable life-threatening infections will be developed soon.
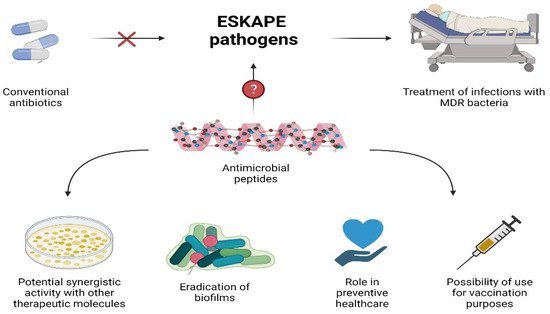
Figure 1. Potential use of AMPs.
References
- Gots, J.S. Production of extracellular penicillin-inactivating substances associated with penicillin resistance in Staphylococcus aureus. Proceedings of the Society for Experimental Biology and Medicine. Soc. Exp. Biol. Med. 1945, 60, 165–168.
- Friedman, N.D.; Temkin, E.; Carmeli, Y. The negative impact of antibiotic resistance. Clinical microbiology and infection: The official publication of the European Society of Clinical Microbiology and Infectious Diseases. Clin. Microbiol. Infect. 2016, 22, 416–422.
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Government of the United Kingdom: London, UK, 2016.
- Yu, G.; Baeder, D.Y.; Regoes, R.R.; Rolff, J. Predicting drug resistance evolution: Insights from antimicrobial peptides and antibiotics. Proc. R. Soc. B Biol. Sci. 2018, 285, 20172687.
- Nicolas, P. Multifunctional host defense peptides: Intracellular-targeting antimicrobial peptides. FEBS J. 2009, 276, 6483–6496.
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016, 6, 194.
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779.
- Wang, E.B.G. Antimicrobial Peptides Discovery, Design and Novel Therapeutic Strategies, 2nd ed.; University of Nebraska Medical Center: Omaha, NE, USA, 2017.
- The Antimicrobial Peptide Database. Available online: https://wangapd3.com/ (accessed on 1 July 2021).
- Ramos-Martín, F.; Annaval, T.; Buchoux, S.; Sarazin, C.; D’Amelio, N. ADAPTABLE: A comprehensive web platform of antimicrobial peptides tailored to the user’s research. Life Sci. Alliance 2019, 2, e201900512.
- Kościuczuk, E.M.; Lisowski, P.; Jarczak, J.; Strzałkowska, N.; Jóźwik, A.; Horbańczuk, J.; Krzyżewski, J.; Zwierzchowski, L.; Bagnicka, E. Cathelicidins: Family of antimicrobial peptides. A review. Mol. Biol. Rep. 2012, 39, 10957–10970.
- Patocka, J.; Nepovimova, E.; Klimova, B.; Wu, Q.; Kuca, K. Antimicrobial Peptides: Amphibian Host Defense Peptides. Curr. Med. Chem. 2018, 25, 5924–5946.
- Masso-Silva, J.A.; Diamond, G. Antimicrobial peptides from fish. Pharmaceuticals 2014, 7, 265–310.
- Wu, Q.; Patočka, J.; Kuča, K. Insect Antimicrobial Peptides, a Mini Review. Toxins 2018, 10, 461.
- Tam, J.P.; Wang, S.; Wong, K.H.; Tan, W.L. Antimicrobial Peptides from Plants. Pharmaceuticals 2015, 8, 711–757.
- Pardoux, É.; Boturyn, D. Antimicrobial Peptides as Probes in Biosensors Detecting Whole Bacteria: A Review. Molecules 2020, 25, 1998.
- Akhtar, M.S.; Imran, M.B.; Nadeem, M.A.; Shahid, A. Antimicrobial peptides as infection imaging agents: Better than radiolabeled antibiotics. Int. J. Pept. 2012, 2012, 965238.
- Mazaheri Tehrani, M.; Erfani, M.; Amirmozafari, N. [(99m) Tc-HYNIC/EDDA]-MccJ25 antimicrobial peptide analog as a potential radiotracer for detection of infection. Chem. Biol. Drug Des. 2021, 97, 904–913.
- Database of Antimicrobial Activity and Structure of Peptides. Available online: https://dbaasp.org/ (accessed on 21 June 2021).
- Balmeh, N.; Mahmoudi, S.; Fard, N.A. Manipulated bio antimicrobial peptides from probiotic bacteria as proposed drugs for COVID-19 disease. Inform. Med. Unlocked 2021, 23, 100515.
- Mahendran, A.S.K.; Lim, Y.S.; Fang, C.-M.; Loh, H.-S.; Le, C.F. The Potential of Antiviral Peptides as COVID-19 Therapeutics. Front. Pharmacol. 2020, 11, 1475.
- Bakovic, A.; Risner, K.; Bhalla, N.; Alem, F.; Chang, T.L.; Weston, W.; Harness, J.A.; Narayanan, A. Brilacidin, a COVID-19 Drug Candidate, Exhibits Potent In Vitro Antiviral Activity Against SARS-CoV-2. bioRxiv 2020.
- Elnagdy, S.; AlKhazindar, M. The Potential of Antimicrobial Peptides as an Antiviral Therapy against COVID-19. ACS Pharmacol. Transl. Sci. 2020, 3, 780–782.
- Liscano, Y.; Oñate-Garzón, J.; Ocampo-Ibáñez, I.D. In Silico Discovery of Antimicrobial Peptides as an Alternative to Control SARS-CoV-2. Molecules 2020, 25, 5535.
More
Information
Subjects:
Medicine, General & Internal
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.5K
Revisions:
2 times
(View History)
Update Date:
23 Sep 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




