
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | François Ferron | + 4468 word(s) | 4468 | 2020-07-22 11:50:53 | | | |
| 2 | Rita Xu | -1852 word(s) | 2616 | 2020-07-27 11:35:40 | | |
Video Upload Options
Arenaviridae is a family of viruses harbouring important emerging pathogens belonging to the Bunyavirales order. Like in other segmented negative strand RNA viruses, the nucleoprotein (NP) is a major actor of the viral life cycle being both (i) the necessary co-factor of the polymerase present in the L protein, and (ii) the last line of defence of the viral genome (vRNA) by physically hiding its presence in the cytoplasm. The NP is also one of the major players interfering with the immune system. Several structural studies of NP have shown that it features two domains: a globular RNA binding domain (NP-core) in its N-terminal and an exonuclease domain (ExoN) in its C-terminal. Further studies have observed that significant conformational changes are necessary for RNA encapsidation. We here present the architecture and latest structural data available on Arenaviridae NP.
1. Definition
Arenaviridae is a family of viruses harbouring important emerging pathogens belonging to the Bunyavirales order. Like in other segmented negative strand RNA viruses, the nucleoprotein (NP) is a major actor of the viral life cycle being both (i) the necessary co-factor of the polymerase present in the L protein, and (ii) the last line of defence of the viral genome (vRNA) by physically hiding its presence in the cytoplasm. The NP is also one of the major players interfering with the immune system. Several structural studies of NP have shown that it features two domains: a globular RNA binding domain (NP-core) in its N-terminal and an exonuclease domain (ExoN) in its C-terminal. Further studies have observed that significant conformational changes are necessary for RNA encapsidation.
2. Introduction
Arenaviruses are zoonotic viruses that cause chronic infections in rodents, which constitute a reservoir of human pathogens across the world. The Arenaviridae family was recently reclassified into the Bunyavirales, a viral order that includes several major human pathogens, including the Rift Valley Fever virus (RVFV), Hantaan virus (HTNV) and Crimean Congo Haemorrhagic Fever virus (CCHFV). Arenaviridae regroups Mammarenavirus, Reptarenavirus, Hartmanivirus, and Antennavirus [1][2]. Mammarenaviruses are further classified into two groups based on geography and phylogeny: the Old World (OW) arenaviruses and the New World (NW) arenaviruses, itself divided into clades A, B, C and D. Several of the Mammarenavirus are responsible for viral haemorrhagic fevers (VHFs) in humans: OW Lassa virus (LASV) and NW clade B: Junin virus (JUNV), Machupo virus (MACV), Guanarito virus (GTOV), Sabia virus (SABV), and Chapare virus (CHAV). The OW prototype Lymphocytic choriomeningitis virus (LCMV) can cause nervous disorders like meningitis and hearing loss [3][4], and is responsible for a large number of miscarriages [5][6] due to neonatal infections and health complications for immune-compromised individuals [7][8][9]. Meanwhile, NW Pichinde virus (PICV) from the prototype Tacaribe Virus (TCRV) complex is non-pathogenic for humans and animals. Recently, OW arenaviruses have been isolated in Asia from mice, shrews and black rats, expanding host variety and geographic distribution of Mammarenavirus [10][11][12][13], and the list of human pathogens [14]. In recent years, repeated LASV outbreaks pointed out the major public health concerns in their regions of endemicity and surroundings [15][16][17][18][19][20] not only due to the severe acute disease and high mortality rates, but also to the long-term sequelae responsible for significant social and economic burdens [21][22]. Finally, the frequency of imported cases in Europe and the USA have increased in the last few years, illustrating the possibility of imported cases of haemorrhagic fever of both OW and NW arenaviruses [23][24][25][26].
Arenaviruses are enveloped viruses containing a segmented negative-sense single-stranded RNA genome (sNSV). Apart from the tri-segmented antennaviruses genus, the RNA genome (vRNA) is comprised of two segments: a large segment (L) of around 7.2 kb and a small segment (S) of around 3.4 kb. Each segment uses an ambisense coding strategy to direct synthesis of two proteins in opposite orientation separated by an intergenic region (IGR). The L segment encodes the large protein L (~ 200 kDa) and a small protein Z (~ 11 kDa) that functions as the matrix protein of the virion. The S segment encodes the multi-functional nucleoprotein (see below) NP (~ 63 kDa) and the glycoprotein precursor (GPC; 75 kDa), that will give after post-translational cleavage, GP1 (40 to 46 kDa), GP2 (35 kDa) and SSP a transmembrane stable signal peptide. The mature glycoprotein (GP) complex on the viral surface is a trimer of heterotrimers composed of GP1/GP2 and SSP. The IGR is thought to fold into secondary structures, which lead to viral messenger RNA (mRNA) transcription termination [27]. The RNA genome (and complementary) is always encapsulated in a polymer of NP forming the ribonucleoprotein complex (RNP). The 5’ and 3’ ends of each segment contain conserved untranslated regions (UTR) that are complementary to each other, forming a panhandle structure at the end of the viral genome [28][29] on which binds the L. The whole (RNP-L) constitutes the replication–transcription complexes (RTC) and NP is a necessary co-factor of L [30]. Like all other Bunyavirales nucleoproteins, NPs are the most abundant viral proteins both in infected cells and virions. They are de facto the main structural and multi-functional component of the viral cycle but, unlike the other nucleoproteins, they are also multi-domain proteins. By coating the vRNA (or anti-genomic), they passively protect the viral genome from degradation, avoid formation of dsRNA between viral RNAs of opposite polarity, and compact the RNA into RNPs. Moreover, they are responsible for generating a significant interference in the transduction pathway signalling cellular infection by recruiting several host proteins. Finally, through the unique presence of an active exonuclease (ExoN), it actively degradates dsRNA, contributing to the silencing of innate immunity and to a yet unclear process seemingly playing a role in replication.
In infected cells, NP binds to vRNA but neither to viral mRNAs nor to cellular mRNA; however, when recombinant NPs are expressed in bacteria, they are observed to bind to host RNAs forming structures reminiscent of the viral RNP structure. This RNP formation suggests that the polymerisation process is tightly regulated in the infected cell.
3. Arenavirus Nucleoprotein Architecture and Structure from Atomic Structures to Observation
3.1. Architecture and Full-Length Structure of NP
Arenavirus NPs are involved in several critical functions for the virus life cycle: transcription/replication, genome/anti genome protection (both passive and active) and genome packaging. These functions are reflected in the structural architecture of NP as a two domain protein, an amino terminal domain (N-terminal) involved in polymerisation and viral RNA protection and a carboxy terminal domain (C-terminal) involved in degrading dsRNA, a marker of viral infection. The latter domain is surrounded by two flexible linkers that impact high resolution structural studies (Figure 1).
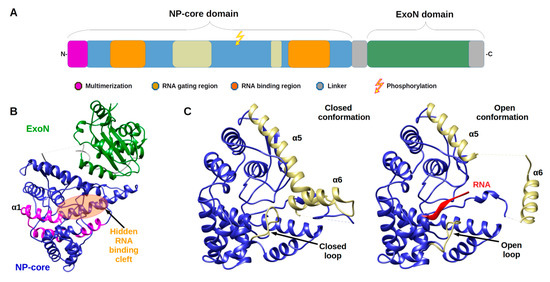
Figure 1. Architecture of Arenaviridae nucleoprotein (NP) and Nucleoprotein structures. (A) Annotated schematic of NP architecture. Colour code of annotation is in caption. (B) Structure of full-length NP (PDB 3MWP) represented in ribbon. NP-core domain is in blue and the exonuclease domain (ExoN) in green, the proposed multimerization arm of the NP-core in pink, the hidden RNA binding cleft highlighted in orange circle. (C) Structure of NP-core domain (blue/ kaki) in open and closed conformation focus on the RNA binding cleft. Left panel shows the RNA binding cleft of the 3MWP structure. Right panel presents the corresponding domain of the structure 3T5Q with RNA (red ribbon). Comparison of these two structures shows that in the absence of RNA, the cavity is closed by the α5 and α6 helix shown in kaki as well as by the loop (residues 234–245) shown also in kaki. The α5 and α6 helix as well as the loop are displaced in the case of the 3T5Q structure, permitting the adsorption of the viral RNA. All structural figures and movies were done using UCSF chimera [31].
The monomeric full-length NP crystal structure of Lassa virus [32][33] presents a two domain protein separated by a flexible linker (of 30 amino acids) uncharacterised in structure due to its intrinsic flexibility (Figure 1b). Both domains are in tight contact with each other, and their respective position is likely due to the crystal packing.
The N-terminal domain is a globular α/β domain composed of 14 α-helices and six β-strands that can undergo significant structural changes (Figure 1c & Movie 1). In all NP structures, the first fifty amino acids (α1, α2) involved in the multimerization mechanism [34] are folded over the core domain, suggesting a mechanism reminiscent of the one described in Phenuiviridae (see below). In the original structures of Qi and collaborator [32], NTP were observed to be trapped within the N-terminal domain. That result suggested a gating mechanism was allowing the access to a potential cavity. Instead, it misled the authors to propose that NP has potential cap-binding activity that could provide the host-derived primers to initiate transcription by the virus polymerase [32]. However, later studies showed that this putative NP cap-binding domain corresponded to the NP RNA binding site and the cap binding domain was identified in the C-terminal part of L protein [35][36][37]. The RNA binding cleft is indeed covered by two helices (α5, α6) and a loop (Figure 1c). A superposition of the structures alone or in complex with a ssRNA clearly presents the change of position of these latter secondary structure elements to create the cleft accommodating the RNA (Figure 1c and Figure S1a and Movie 1). In the RNA bound form, several positions of the helix (α6) are observed showing a large degree of liberty in its positioning (7 different positions are observed) [35].
The binding of the RNA and the opening of the cleft impact the secondary structure elements forcing the repositioning of the C-terminal domain to a hypothetical and not yet characterised position [35] (Figure S1b).
Arenavirus NP, unlike other RNA negative stranded viruses (NSV) nucleoproteins, has acquired a 3′-5′ exonuclease domain specific to dsRNA in its C-terminal, exhibiting a type I interferon (IFN-I)-counteracting activity [38][39][40]. Moreover, this domain does not seem to be required for replication or transcription of the viral genome. This assertion, however, needs to be carefully evaluated as PICV, LASV and LCMV with a mutant NP lacking the 3′-5′ exonuclease activity had either not rescuable, impaired replication or a significant decrease in fitness during its replication [38][41][42]. Moreover, a recent in vitro study has shown a removing capability of mismatched nucleotide, which could be a first step in an editing process [43]. The ExoN belongs to the DEDDh family, that process their substrate through a two-metal ion catalytic mechanism [44], the two ions being coordinated by the residues of the motif. A sequence analysis of the ExoN domain of Arenaviridae shows the evolution of the catalytic motifs DEDDh to DEEDh (Figure 2 and Figure S2), an observation that added to the topology and the in vitro activity conservation with the ExoN domaine of Coronaviridae Nsp14, suggesting a common origin of the Coronaviridae nsp14 and Arenaviridae NP ExoNs [43][45]. This domain has a canonical fold of the DEDDh family of 3′- 5′ exoribonucleases, consisting of two β-sheets (with six mixed strands and two anti-parallel strands) and eight α-helices connected by a series of loops. These secondary structure elements are arranged to form the central β-sheet sandwiched by three α-helices on one side and seven α-helices on the opposite side and structured by a Zn binding site highly conserved in arenaviruses (Figure 2a). The reported structures often present one metallic ion in the catalytic site [46][47][48][49][50], the second ion, allowing the catalytic reaction, comes dynamically with the RNA substrate (Figure 2a zoom) [50]. Two flexible regions are clearly defined within the structure. The ‘basic loop’ sometimes structures itself as two anti-parallel strands (residues 514–526) above the active site and the C-terminal arm (residues 549–570). It is worth noting that in the full length structure, the latter region was folded over, between the NP-core and ExoN, while in ExoN domain structure, observation of crystal packing reveals that the C-terminal extends away from the domain core towards the back of the next ExoN core. Sequence alignment analysis shows a conserved hydrophobic patch at the C-terminal (Figure S3). This suggests that the C-terminal could also be involved in stabilizing the ExoN domain within the polymer assembly and packing (Figure S3).
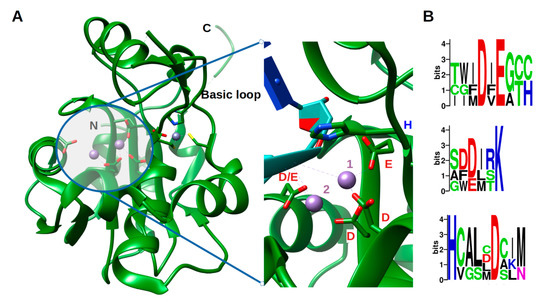
Figure 2. Structure of the exonuclease domain (ExoN) and conservation of the catalytic site. (A) Annotated structure of the ExoN domain represented in ribbon with ions Mn2+ in purple and Zn2+ in grey and zoom on the catalytic residues DEDDh shown in sticks with catalytic ions and 3′ end of double-stranded (ds)RNA substrate (cyan) (PDB). Metallic ion Mn2+ are marked as 1 and 2, 1 being the ion that is always observed and 2 the ion dynamically brought by the RNA. (B) Weblogo [51] of the DEDDh catalytic site through Arenaviridae.
3.2. From Filament to Polymer Assembly
The arenavirus RNP structural data are sparse, yet in the light of recent data, extremely informative on its general assembly [52]. The low-resolution EM structure of PICV RNP shows that it is mostly formed by a flexible structure composed of NP monomers assembled linearly, and forming a filament. This filament appears to fold progressively through a number of intermediate helical structures, that reveal an increasing number of NPs associated with each turn of the helix. They range from a fragile configuration of two to three NPs per turn to a more stable fibre-like structure in which the number of NPs could not be resolved [52]. Furthermore, additional packaging levels were observed with the presence of supercoiled structures [52] forming fibres with a diameter of 15 nm. From more recent EM studies, these trimeric assemblies were rediscovered and combined with high resolution crystallographic data [33]. Moreover, from bacterially- expressed and purified NP of Mopeia virus (MOPV), we have recently measured the full length MOPV N protein by negative stain Transmission Electron Microscopy (TEM). This offers some new results concerning the multimerization shown in Figure 3 (preliminary data and unpublished observation). These RNP particles were observed with a diameter 14 ± 1 nm as well as symmetric circular heptamers of the same diameter (Figure 3a), a result consistent with the original measurement of PICV RNP purified from the virus (Figure 1c of [52]). Unfortunately, near atomic cryo-microscopy data on these polymers are missing to place the packaging position of both domains in the RNP. However, using the observed particles, a low resolution 3D particle reconstruction using the EMAN2 pipeline procedure [53] gives the result shown in Figure 3b together with the corresponding Fourier Shell Correlation (FSC) coefficient in function of the spatial frequencies in Å−1 units and particle classes used in the refinement. This multimerization trend is reminiscent of the original observation [52] but differs from the trimeric NP complexes previously observed [33][35], and additional observations are needed in order to understand the multimerization mechanism of the NP. The various multimerization fashions reported may concern the RNP structure, or the NP in solution, or during the intracellular stage of NP accumulation, or interaction with cellular cofactors recruited by the NP before assembly into RNP polymers [35].
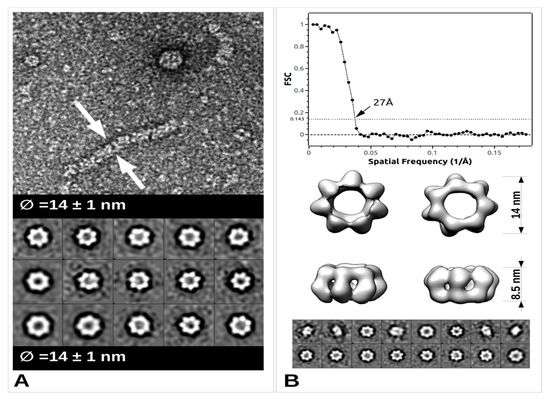
Figure 3. TEM images and data from freshly purified Mopeia virus (MOPV) NP protein. (A) Ribonucleoprotein complex (RNP) particle and several classses showing heptamer organisation (0.05 mg/mL). A 5 μL drop was applied to a freshly deposited and glow-discharged formvar-carbon-coated grid (Copper 300). The grid was stained with Nano-W® (Nanoprobes) and transferred into a Tecnai 120 kV Electron Microscope. A total of 100 raw images were recorded with an EAGLE 2k × 2k CCD camera. Images were under-focused at 1–2 μm with a final resolution of 2.8 Å/pix. Boxing, classification, initial model calculation, as well as refinement for 3D reconstruction, was done with the EMAN2 pipeline [53]. Arrows indicate the sides of the measured object. (B) Top: Graph of the Fourier Shell Correlation (FSC) coefficient in function of the spatial frequencies in Å−1, arrow indicating the maximum resolution; Central: 3D reconstruction at 27Å resolution with below corresponding particle classes used (1224 particles).
References
- Maes, P.; Alkhovsky, S.V.; Bào, Y.; Beer, M.; Birkhead, M.; Briese, T.; Buchmeier, M.J.; Calisher, C.H.; Charrel, R.N.; Choi, I.R.; et al. Taxonomy of the family Arenaviridae and the order Bunyavirales: Update 2018. Arch. Virol. 2018, 163, 2295–2310, doi:10.1007/s00705-018-3843-5.
- Abudurexiti, A.; Adkins, S.; Alioto, D.; Alkhovsky, S.V.; Avšič-Županc, T.; Ballinger, M.J.; Bente, D.A.; Beer, M.; Bergeron, É.; Blair, C.D.; et al. Taxonomy of the order Bunyavirales: Update 2019. Arch. Virol. 2019, 164, 1949–1965, doi:10.1007/s00705-019-04253-6.
- Hirsch, E. Sensorineural deafness and labyrinth damage due to lymphocytic choriomeningitis. Report of a case; Arch. Otolaryngol. 1976, 102, 499–500, doi:10.1001/archotol.1976.00780130093013.
- Ormay, I.; Kovács, P. [Lymphocytic choriomeningitis causing unilateral deafness]. Orv. Hetil. 1989, 130, 789–791.
- Jamieson, D.J.; Kourtis, A.P.; Bell, M.; Rasmussen, S.A. Lymphocytic choriomeningitis virus: An emerging obstetric pathogen? Am. J. Obs. Gynecol. 2006, 194, 1532–1536, doi:10.1016/j.ajog.2005.11.040.
- Barton, L.L.; Mets, M.B.; Beauchamp, C.L. Lymphocytic choriomeningitis virus: Emerging fetal teratogen. Am. J. Obstet. Gynecol. 2002, 187, 1715–1716, doi:10.1067/mob.2002.126297.
- Mets, M.B.; Barton, L.L.; Khan, A.S.; Ksiazek, T.G. Lymphocytic choriomeningitis virus: An underdiag-nosed cause of congenital chorioretinitis. Am. J. Ophthalmol. 2000, 130, 209–215, doi:10.1016/S0002-9394(00)00570-5.
- Brézin, A.P.; Thulliez, P.; Cisneros, B.; Mets, M.B.; Saron, M.-F. Lymphocytic choriomeningitis virus cho-rioretinitis mimicking ocular toxoplasmosis in two otherwise normal children. Am. J. Ophthalmol. 2000, 130, 245–247, doi:10.1016/S0002-9394(00)00563-8.
- Fischer, S.A.; Graham, M.B.; Kuehnert, M.J.; Kotton, C.N.; Srinivasan, A.; Marty, F.M.; Comer, J.A.; Guarner, J.; Paddock, C.D.; DeMeo, D.L.; et al. Transmission of lymphocytic choriomeningitis virus by organ transplantation. N. Engl. J. Med. 2006, 354, 2235–2249, doi:10.1056/NEJMoa053240.
- Li, K.; Lin, X.D.; Li, M.H.; Wang, M.R.; Sun, X.Y.; Zhang, Y.Z. [Genomic analysis of Wenzhou virus in rodents from Zhejiang province]. Zhonghua Liu Xing Bing Xue Za Zhi 2017, 38, 384–387, doi:10.3760/cma.j.issn.0254-6450.2017.03.022.
- Blasdell, K.R.; Becker, S.D.; Hurst, J.; Begon, M.; Bennett, M. Host range and genetic diversity of arena-viruses in rodents, United Kingdom. Emerg. Infect. Dis. 2008, 14, 1455–1458, doi:10.3201/eid1409.080209.
- Li, K.; Lin, X.-D.; Wang, W.; Shi, M.; Guo, W.-P.; Zhang, X.-H.; Xing, J.-G.; He, J.-R.; Wang, K.; Li, M.-H.; et al. Isolation and characterization of a novel arenavirus harbored by Rodents and Shrews in Zhejiang province, China. Virology 2015, 476, 37–42, doi:10.1016/j.virol.2014.11.026.
- Tan, Z.; Yu, H.; Xu, L.; Zhao, Z.; Zhang, P.; Qu, Y.; He, B.; Tu, C. Virome profiling of rodents in Xinjiang Uygur Autonomous Region, China: Isolation and characterization of a new strain of Wenzhou virus. Vi-rology 2019, 529, 122–134, doi:10.1016/j.virol.2019.01.010.
- Blasdell, K.R.; Duong, V.; Eloit, M.; Chretien, F.; Ly, S.; Hul, V.; Deubel, V.; Morand, S.; Buchy, P. Evi-dence of human infection by a new mammarenavirus endemic to Southeastern Asia. eLife 2016, 5, e13135, doi:10.7554/eLife.13135.
- WHO. Lassa Fever—Benin, Togo and Burkina Faso. Available online: http://www.who.int/csr/don/10-march-2017-lassa-fever-benin-togo-burkina-faso/en/ (accessed on Jun 7, 2020).
- WHO. Lassa Fever—Liberia. Available online: http://www.who.int/csr/don/18-may-2016-lassa-fever-liberia/en/ (accessed on 7 June 2020).
- WHO. Lassa Fever—Nigeria. Available online: http://www.who.int/csr/don/20-february-2020-lassa-fever-nigeria/en/ (accessed on 7 June 2020).
- WHO. Lassa Fever—Nigeria. Available online: http://www.who.int/csr/don/14-february-2019-lassa-fever-nigeria/en/ (accessed on 7 June 2020).
- WHO. Lassa Fever—Nigeria. Available online: http://www.who.int/csr/don/20-april-2018-lassa-fever-nigeria/en/ (accessed on 7 June 2020).
- Shehu, N.Y.; Gomerep, S.S.; Isa, S.E.; Iraoyah, K.O.; Mafuka, J.; Bitrus, N.; Dachom, M.C.; Ogwuche, J.E.; Onukak, A.E.; Onyedibe, K.I.; et al. Lassa Fever 2016 Outbreak in Plateau State, Nigeria-The Chang-ing Epidemiology and Clinical Presentation. Front. Public Health 2018, 6, 232, doi:10.3389/fpubh.2018.00232.
- Mateer, E.J.; Huang, C.; Shehu, N.Y.; Paessler, S. Lassa fever-induced sensorineural hearing loss: A ne-glected public health and social burden. PLoS Negl. Trop. Dis. 2018, 12, e0006187, doi:10.1371/journal.pntd.0006187.
- Mehand, M.S.; Al-Shorbaji, F.; Millett, P.; Murgue, B. The WHO R&D Blueprint: 2018 review of emerging infectious diseases requiring urgent research and development efforts. Antivir. Res. 2018, 159, 63–67, doi:10.1016/j.antiviral.2018.09.009.
- WHO. Lassa Fever—United States of America. Available online: https://www.who.int/csr/don/28-may-2015-lassa-fever-usa/en/ (accessed on 7 June 2020).
- WHO. Lassa Fever—Germany. Available online: http://www.who.int/csr/don/27-april-2016-lassa-fever-germany/en/ (accessed on 7 June 2020).
- WHO. Lassa Fever—Sweden. Available online: http://www.who.int/csr/don/8-april-2016-lassa-fever-sweden/en/ (accessed on 7 June 2020).
- Veliziotis, I.; Roman, A.; Martiny, D.; Schuldt, G.; Claus, M.; Dauby, N.; Van den Wijngaert, S.; Martin, C.; Nasreddine, R.; Perandones, C.; et al. Clinical Management of Argentine Hemorrhagic Fever using Ribavirin and Favipiravir, Belgium, 2020. Emerg. Infect. Dis. 2020, 26, 1562–1566, doi:10.3201/eid2607.200275.
- Pinschewer, D.D.; Perez, M.; de la Torre, J.C. Dual role of the lymphocytic choriomeningitis virus inter-genic region in transcription termination and virus propagation. J. Virol. 2005, 79, 4519–4526, doi:10.1128/JVI.79.7.4519-4526.2005.
- Buchmeier, M.J.; de la Torre, J.-C.; Peters, C.J.; Torre, J.D. Arenaviridae: The Viruses and Their Replica-tion. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; Volume II, pp. 1791–1827.
- Perez, M.; Craven, R.C.; de la Torre, J.C. The small RING finger protein Z drives arenavirus budding: Implications for antiviral strategies. Proc. Natl. Acad. Sci. USA 2003, 100, 12978–12983, doi:10.1073/pnas.2133782100.
- Kranzusch, P.J.; Schenk, A.D.; Rahmeh, A.A.; Radoshitzky, S.R.; Bavari, S.; Walz, T.; Whelan, S.P.J. As-sembly of a functional Machupo virus polymerase complex. Proc. Natl. Acad. Sci. USA 2010, 107, 20069–20074, doi:10.1073/pnas.1007152107.
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612, doi:10.1002/jcc.20084.
- Qi, X.; Lan, S.; Wang, W.; Schelde, L.M.; Dong, H.; Wallat, G.D.; Ly, H.; Liang, Y.; Dong, C. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature 2010, 468, 779–783, doi:10.1038/nature09605.
- Brunotte, L.; Kerber, R.; Shang, W.; Hauer, F.; Hass, M.; Gabriel, M.; Lelke, M.; Busch, C.; Stark, H.; Svergun, D.I.; et al. Structure of the Lassa virus nucleoprotein revealed by X-ray crystallography, small-angle X-ray scattering, and electron microscopy. J. Biol. Chem. 2011, 286, 38748–38756, doi:10.1074/jbc.M111.278838.
- Ortiz-Riaño, E.; Cheng, B.Y.H.; de la Torre, J.C.; Martínez-Sobrido, L. Self-association of lymphocytic choriomeningitis virus nucleoprotein is mediated by its N-terminal region and is not required for its anti-interferon function. J. Virol. 2012, 86, 3307–3317, doi:10.1128/JVI.05503-11.
- Hastie, K.M.; Liu, T.; Li, S.; King, L.B.; Ngo, N.; Zandonatti, M.A.; Woods, V.L.; de la Torre, J.C.; Saphire, E.O. Crystal structure of the Lassa virus nucleoprotein-RNA complex reveals a gating mechanism for RNA binding. Proc. Natl. Acad. Sci. USA 2011, 108, 19365–19370, doi:10.1073/pnas.1108515108.
- Rosenthal, M.; Gogrefe, N.; Vogel, D.; Reguera, J.; Rauschenberger, B.; Cusack, S.; Günther, S.; Reindl, S. Structural insights into reptarenavirus cap-snatching machinery. PLoS Pathog. 2017, 13, e1006400, doi:10.1371/journal.ppat.1006400.
- Peng, R.; Xu, X.; Jing, J.; Wang, M.; Peng, Q.; Liu, S.; Wu, Y.; Bao, X.; Wang, P.; Qi, J.; et al. Structural insight into arenavirus replication machinery. Nature 2020, 579, 615–619, doi:10.1038/s41586-020-2114-2.
- Martínez-Sobrido, L.; Emonet, S.; Giannakas, P.; Cubitt, B.; García-Sastre, A.; de la Torre, J.C. Identifica-tion of amino acid residues critical for the anti-interferon activity of the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J. Virol. 2009, 83, 11330–11340, doi:10.1128/JVI.00763-09.
- Martínez-Sobrido, L.; Giannakas, P.; Cubitt, B.; García-Sastre, A.; de la Torre, J.C. Differential inhibition of type I interferon induction by arenavirus nucleoproteins. J. Virol. 2007, 81, 12696–12703, doi:10.1128/JVI.00882-07.
- Martínez-Sobrido, L.; Zúñiga, E.I.; Rosario, D.; García-Sastre, A.; de la Torre, J.C. Inhibition of the type I interferon response by the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis vi-rus. J. Virol. 2006, 80, 9192–9199, doi:10.1128/JVI.00555-06.
- Huang, Q.; Shao, J.; Lan, S.; Zhou, Y.; Xing, J.; Dong, C.; Liang, Y.; Ly, H. In vitro and in vivo character-izations of pichinde viral nucleoprotein exoribonuclease functions. J. Virol. 2015, 89, 6595–6607, doi:10.1128/JVI.00009-15.
- Carnec, X.; Baize, S.; Reynard, S.; Diancourt, L.; Caro, V.; Tordo, N.; Bouloy, M. Lassa virus nucleopro-tein mutants generated by reverse genetics induce a robust type I interferon response in human dendritic cells and macrophages. J. Virol. 2011, 85, 12093–12097, doi:10.1128/JVI.00429-11.
- Yekwa, E.; Aphibanthammakit, C.; Carnec, X.; Coutard, B.; Picard, C.; Canard, B.; Baize, S.; Ferron, F. Arenaviridae exoribonuclease presents genomic RNA edition capacity. BioRxiv 2019, 541698, doi: https://doi.org/10.1101/541698.
- Steitz, T.A.; Steitz, J.A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. USA 1993, 90, 6498–6502, doi:10.1073/pnas.90.14.6498.
- Emonet, S.E.; Urata, S.; de la Torre, J.C. Arenavirus reverse genetics: New approaches for the investiga-tion of arenavirus biology and development of antiviral strategies. Virology 2011, 411, 416–425, doi:10.1016/j.virol.2011.01.013.
- Zhang, Y.; Li, L.; Liu, X.; Dong, S.; Wang, W.; Huo, T.; Guo, Y.; Rao, Z.; Yang, C. Crystal structure of Junin virus nucleoprotein. J. Gen. Virol. 2013, 94, 2175–2183, doi:10.1099/vir.0.055053-0.
- West, B.R.; Hastie, K.M.; Saphire, E.O. Structure of the LCMV nucleoprotein provides a template for un-derstanding arenavirus replication and immunosuppression. Acta Cryst. D Biol. Cryst. 2014, 70, 1764–1769, doi:10.1107/S1399004714007883.
- Hastie, K.M.; King, L.B.; Zandonatti, M.A.; Saphire, E.O. Structural basis for the dsRNA specificity of the Lassa virus NP exonuclease. PLoS ONE 2012, 7, e44211, doi:10.1371/journal.pone.0044211.
- Yekwa, E.; Khourieh, J.; Canard, B.; Papageorgiou, N.; Ferron, F. Activity inhibition and crystal poly-morphism induced by active-site metal swapping. Acta Cryst. D Struct. Biol. 2017, 73, 641–649, doi:10.1107/S205979831700866X.
- Jiang, X.; Huang, Q.; Wang, W.; Dong, H.; Ly, H.; Liang, Y.; Dong, C. Structures of arenaviral nucleo-proteins with triphosphate dsRNA reveal a unique mechanism of immune suppression. J. Biol. Chem. 2013, 288, 16949–16959, doi:10.1074/jbc.M112.420521.
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190, doi:10.1101/gr.849004.
- Young, P.R.; Howard, C.R. Fine structure analysis of Pichinde virus nucleocapsids. J. Gen. Virol. 1983, 64 Pt 4, 833–842, doi:10.1099/0022-1317-64-4-833.
- Tang, G.; Peng, L.; Baldwin, P.R.; Mann, D.S.; Jiang, W.; Rees, I.; Ludtke, S.J. EMAN2: An extensible im-age processing suite for electron microscopy. J. Struct. Biol. 2007, 157, 38–46, doi:10.1016/j.jsb.2006.05.009.
- Raymond, D.D.; Piper, M.E.; Gerrard, S.R.; Skiniotis, G.; Smith, J.L. Phleboviruses encapsidate their ge-nomes by sequestering RNA bases. Proc. Natl. Acad. Sci. USA 2012, 109, 19208–19213, doi:10.1073/pnas.1213553109.
- Ferron, F.; Li, Z.; Danek, E.I.; Luo, D.; Wong, Y.; Coutard, B.; Lantez, V.; Charrel, R.; Canard, B.; Walz, T.; et al. The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its as-sembly into ribonucleoprotein complexes. PLoS Pathog. 2011, 7, e1002030, doi:10.1371/journal.ppat.1002030.
- Baklouti, A.; Goulet, A.; Lichière, J.; Canard, B.; Charrel, R.N.; Ferron, F.; Coutard, B.; Papageorgiou, N. Toscana virus nucleoprotein oligomer organization observed in solution. Acta Cryst. D Struct. Biol. 2017, 73, 650–659, doi:10.1107/S2059798317008774.




