
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Lin gang He | + 1565 word(s) | 1565 | 2021-09-13 04:24:58 |
Video Upload Options
Autophagy with endoplasmic reticulum (ER) as a specific substrate is called ER-phagy or reticulophagy. It occurs both under physiological conditions at the basal level, and when cells are insulted by starvation, UPR, toxin stimulation, and many other internal or external environmental changes, to achieve cell homeostasis by removing damaged or excess ER.
1. Introduction
Selective autophagy is a class of autophagic pathways that targets specific substrates for degradation. Depending on the substrates, selective autophagy can be classified into mitophagy, lysophagy, aggrephagy, pexophagy, nucleophagy, xenophagy, and ER-phagy (endoplasmic reticulum-selective autophagy), etc[1]. Receptor proteins determine the selectivity of autophagic substrates.
ER-phagy, which is also called reticulophagy, refers to the selective autophagy of the ER. Cells use it to selectively clean up damaged ER subdomains and abnormally accumulated ER luminal proteins to maintain homeostasis. With the discovery of the first ER-phagy receptor in 2015, this field has really entered the study at the molecular level and significant mechanisms have been unveiled. At the same time, a close relationship between ER-phagy and human diseases, such as neurodegenerative diseases, cancers, and metabolic diseases, has also been found.
2. ER-Phagy
The phenomenon of ER being engulfed by the double-membrane vesicle, autophagosomes, was first reported in 1973[2]. Phenobarbital treatment can significantly induce the formation of inner membrane structure within hepatocytes. After removing the drug, the slippery ER is preferentially degraded through the autophagy pathway, which puts forward the concept of ER-phagy[2]. Later, it was found that rapamycin, an mTOR inhibitor that mimics starvation conditions, can also induce the degradation of ER in yeast, which is dependent on the autophagy machinery[3]. Interestingly, typical UPR inducers such as DTT (dithiothreitol, a reducing agent which breaks disulfide bonds within proteins and maintains the sulfhydryl group in a reduced state, causing serve accumulation of unfolded proteins within ER) and thapsigargin (Tg, a sarcoplasmic reticulum/ ER Ca 2+ -ATPase inhibitor) induce the formation a special ER structure which is called ER whorl[4][5]. In mammalian cells, Fang Xu et al. found that Tg induces ER whorls, which is dependent on PERK activation and COPII machinery[5]. Sebastián Bernales et al. found that the treatment of yeast cells with DTT leads to the dramatic increase of ER volume and also induces the formation of ER whorls[6]. Late on, the same lab found that these ER whorls can be delivered into the vacuole independent of the core macro-autophagy machinery[4]. Therefore, it was being debated whether vacuolar ER degradation is mediated by the autophagic pathway or not. The big breakthrough of this field happened in 2015 when Keisuke Mochida et al. and Aliaksandr Khaminets et al. reported the first ER-phagy receptors in yeast cells and mammalian cells, respectively[7][8]. The discovery of receptor proteins that target ER into autophagosomes gave a clear answer to the existence of selective autophagic degradation of ER.
ER-phagy is a multistep process that needs specific receptors and core autophagic machinery to promote the degradation of ER components. Under the induction of ER stress, such as UPR, protein aggregation, nutritional deficiency, and the destruction of ER structure, the ER components which need to be degraded can be recognized and “labeled” by specific ER-phagy receptors (Figure 1). At the same time, the cells activate the autophagosome initiation complex mainly by inhibiting mTOR or direct phosphorylation of Atg1/ULK1 (Serine/threonine-protein kinase ULK1, Atg1’s homolog in mammals) by AMPK to initiate the assembly of the isolation membrane. Ubiquitin-like proteins, Atg8/LC3/GABARAP, will be recruited to the growing isolation membrane to help membrane expansion. Meanwhile, Atg8/LC3/GABARAP proteins can identify and directly bind ER-phagy receptors. In this way, the ER subdomains to be degraded as well as the ER-phagy receptors are targeted into the autophagosomes as the isolation membranes gradually grow and seal. After that, the ER-containing autophagosomes fuse with the lysosome to degrade the substrate[9][10].
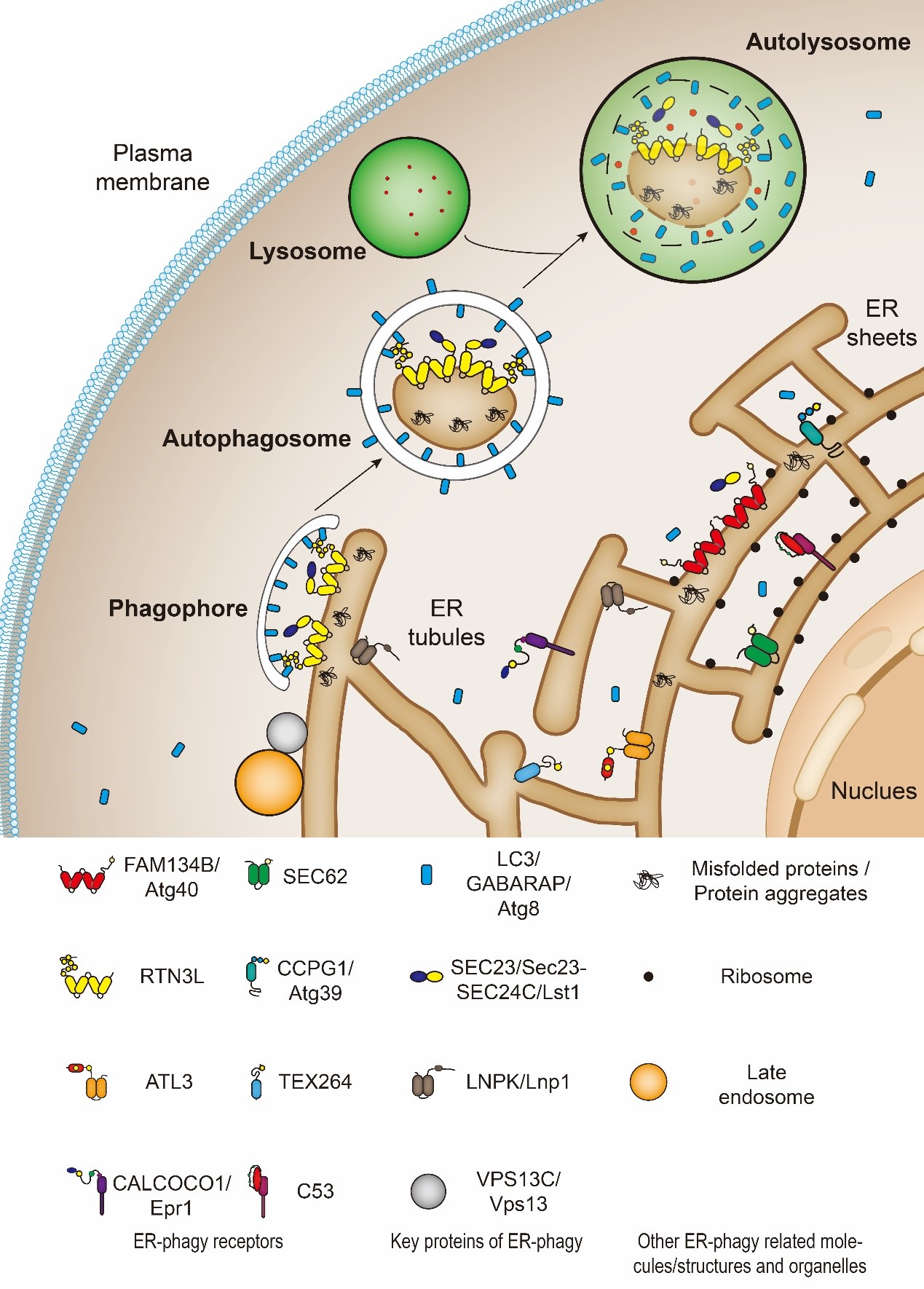
Figure 1. The catabolic process of endoplasmic reticulum-selective autophagy (ER-phagy). FAM134B, Atg40, SEC62, RTN3L, CCPG1, Atg39, ATL3, TEX264, CALCOCO1, Epr1 and C53 are ER-phagy receptors. SEC23/Sec23-SEC24C/Lst1, LNPK/LNP1 and VPS13C/Vps13 are cofactors which function together with ER-phagy receptors.
Like macro-autophagy, ER-phagy is also regulated by signal transduction[7][8][11]. Different stimuli can induce autophagy in different regions of ER, and the receptors and regulatory factors involved are different. And different receptors may participate in ER-phagy at the same time to preserve cell homeostasis[12]. What’s more, in the process of ER-phagy, the expression level, localization, and post-translational modifications (phosphorylation, ubiquitination, etc.) of receptors and other participating proteins are also tightly regulated. Some post-translational modifications are crucial for ER-phagy[13][14][15][16].
3. Key Molecules of ER-Phagy (Receptors, Cofactors, etc.)
Like other selective autophagy, the occurrence of ER-phagy requires specific receptors. In principle, these receptors are either directly or indirectly located on the ER membrane and harbor AIM (Atg8 interaction motif)/LIR (LC3 interaction region)/GIM (GABARAP interaction motif), by which these receptors bind to Atg8/LC3/GABARAP and bridge ER to the autophagosomes.
So far, eleven ER-phagy receptors have been identified from different species. They are either ER-membrane-anchored proteins or ER-membrane-associated proteins. Among them, six in mammals (FAM134B, RTN3L, TEX264, ATL3, SEC62, and CCPG1) and two in budding yeasts (Atg40 and Atg39) are directly anchored on the ER membrane by their reticulon domains or transmembrane domains. The rest identified receptors (CALCOCO1, C53, and Epr1) are located in the ER by binding to ER membrane resident proteins.
In mammalian cells, FAM134B is located on the sheet ER, thus, mediating sheet ER degradation[7]; RTN3L, TEX264, and ATL3 are located on the tubular ER, therefore, responsible for tubular ER clearance[12][17][18][19]; SEC62 is required for recov-ER-phagy, which refers to the ER-phagy induced to restore the homeostasis from ER-stress[20]; CCPG1 involves in maintaining the protein homeostasis of ER[11]; the most-recently discovered ER-phagy receptor, CALCOCO1, is very special because it is the first soluble and non-ER resident protein which mediates the self-bite of ER[21][22]. In budding yeast cells, Atg39 locates on the perinuclear ER (pnER); Atg40 is mainly present on the cytoplasmic ER (cytoER) and cortical ER (cER)[8][23][24]. Epr1 is another soluble ER-phagy receptor protein found in fission yeast and functions like mammalian CALCOCO1[25][26]. In addition, C53 is a soluble ER-phagy receptor found in both plant and mammalian cells[27]. The discovery of these diverse receptors has greatly promoted the understanding of ER-phagy and enriched the entire field of autophagy.
Besides ER-phagy receptors, other proteins such as SEC23/Sec23-SEC24C/Lst1, Lnp1 and Vps13 are also involved in ER-phagy. They work as cofactors of ER-phagy. For example, Yixian Cui et al. recently found that Lst1/SEC24C, a cargo adaptor, assists receptor proteins, Atg40/FAM134B/RTN3L, to mediate ER-phagy[28][29]. They can be transported to the lysosomes along with the autophagosomes during ER-phagy[28]. In addition, Lst1/SEC24C binds with Sec23/SEC23 to form ER-phagy sites (ERPHS), which is distinct from specialized ER exit sites (ERES) where COPII vesicles on the secretory pathway bud and transport to the Golgi[28][29]. Another cofactor is Lnp1. Lnp1 is an ER membrane protein which locates at the three-way junction of the ER[30]. It has been reported that Lnp1 also involves Atg40-mediated-ER-phagy[28][31]. Lnp1 is required for the recruitment of Atg40 to the ERPHS and its co-localization with Atg11[28][31]. And like Lst1 and Lnp1, researchers have found that yeast Vps13, which locates at numerous membrane contact sites, functions together with Atg40 in cortical ER-phagy[32]. It is responsible for targeting the ER fragments into the autophagosomes[32].
4. The Diseases Relevance of ER-Phagy
As a crucial basic biological process, ER-phagy requires to be highly controlled, and insufficient or overactivated ER-phagy could be both pathogenic. Many associations between ER-phagy and human diseases have already been reported, such as neurodegenerative disorders, cancer, metabolic diseases, viral infection, etc.
Neurodegenerative disorders can be characterized by incorrect folding of certain proteins or changes in the intracellular stress response[33]. As an important way to maintain the homeostasis of ER, the main site of protein folding and modification in a cell, ER-phagy plays an important role in clearing misfolded proteins and dealing with intracellular stress. Studies have shown that dysregulated ER-phagy causes a variety of neurodegenerative diseases.
The role of ER-phagy in tumorigenesis is complex. On the one hand, ER-phagy can attenuate excessive ER stress and aid the proliferation and survival of cancer cells. On the other hand, ER-phagy may also induce the death of cancer cells. Its function in tumorigenesis is highly contextual, and likely dependent on the cancer type, progression stage as well as microenvironments.
To maintain proteostasis, multiple degradation pathways, including proteasomal and lysosomal degradation pathways, work together to clear aberrant protein in cells. ER-phagy can target misfolded proteins accumulated in ER to the lysosomes for degradation. When ER-phagy is deficient chronically, it may cause metabolic diseases.
Viruses invade host cells through a variety of methods to avoid cell detection and immune response. To protect from viral infection, cells have evolved multiple strategies, including ER-phagy. However, in most cases, viruses actively impede ER-phagy and other immune responses of cells to fulfill their proliferative needs. In these processes, pathogens usually impair the normal function of ER-phagy receptors and ER-phagy related molecules, then hijack them.
5. Conclusion
Although a lot is known about ER-phagy and its role in physiological and pathological conditions, there are still lots of unknowns to explore. In a similar line to other kinds of selective autophagy, such as xenophagy and mitophagy, efforts to uncover the mechanism of ER-phagy will benefit human health greatly.
References
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)1. Autophagy 2021, 17, 1-382, doi:10.1080/15548627.2020.1797280.
- Bolender , R.P.; Weibel , E.R. A morphometric study of the removal of phenobarbital-induced membranes from hepatocytes after cessation of threatment. Journal of Cell Biology 1973, 56, 746-761, doi:10.1083/jcb.56.3.746
- Hamasaki, M.; Noda, T.; Baba, M.; Ohsumi, Y. Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic 2005, 6, 56-65, doi:10.1111/j.1600-0854.2004.00245.x.
- Schuck, S.; Gallagher, C.M.; Walter, P. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J Cell Sci 2014, 127, 4078-4088, doi:10.1242/jcs.154716.
- Xu, F.; Du, W.; Zou, Q.; Wang, Y.; Zhang, X.; Xing, X.; Li, Y.; Zhang, D.; Wang, H.; Zhang, W.; et al. COPII mitigates ER stress by promoting formation of ER whorls. Cell Res 2021, 31, 141-156, doi:10.1038/s41422-020-00416-2.
- Bernales, S.; McDonald, K.L.; Walter, P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol 2006, 4, e423, doi:10.1371/journal.pbio.0040423.
- Khaminets, A.; Heinrich, T.; Mari, M.; Grumati, P.; Huebner, A.K.; Akutsu, M.; Liebmann, L.; Stolz, A.; Nietzsche, S.; Koch, N.; et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 2015, 522, 354-358, doi:10.1038/nature14498.
- Mochida, K.; Oikawa, Y.; Kimura, Y.; Kirisako, H.; Hirano, H.; Ohsumi, Y.; Nakatogawa, H. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 2015, 522, 359-362, doi:10.1038/nature14506.
- Wilkinson, S. Emerging Principles of Selective ER Autophagy. J Mol Biol 2020, 432, 185-205, doi:10.1016/j.jmb.2019.05.012.
- Grumati, P.; Dikic, I.; Stolz, A. ER-phagy at a glance. J Cell Sci 2018, 131, doi:10.1242/jcs.217364.
- Smith, M.D.; Harley, M.E.; Kemp, A.J.; Wills, J.; Lee, M.; Arends, M.; von Kriegsheim, A.; Behrends, C.; Wilkinson, S. CCPG1 Is a Non-canonical Autophagy Cargo Receptor Essential for ER-Phagy and Pancreatic ER Proteostasis. Dev Cell 2018, 44, 217-232 e211, doi:10.1016/j.devcel.2017.11.024.
- An, H.; Ordureau, A.; Paulo, J.A.; Shoemaker, C.J.; Denic, V.; Harper, J.W. TEX264 Is an Endoplasmic Reticulum-Resident ATG8-Interacting Protein Critical for ER Remodeling during Nutrient Stress. Mol Cell 2019, 74, 891-908 e810, doi:10.1016/j.molcel.2019.03.034.
- Wild, P.; Farhan, H.; McEwan, D.G.; Wagner, S.; Rogov, V.V.; Brady, N.R.; Richter, B.; Korac, J.; Waidmann, O.; Choudhary, C.; et al. Phosphorylation of the Autophagy Receptor Optineurin Restricts Salmonella Growth. 2011, 333, 228-233, doi:10.1126/science.1205405 %J Science.
- Yang, H.; Ni, H.M.; Guo, F.; Ding, Y.; Shi, Y.H.; Lahiri, P.; Frohlich, L.F.; Rulicke, T.; Smole, C.; Schmidt, V.C.; et al. Sequestosome 1/p62 Protein Is Associated with Autophagic Removal of Excess Hepatic Endoplasmic Reticulum in Mice. J Biol Chem 2016, 291, 18663-18674, doi:10.1074/jbc.M116.739821.
- Ji, C.H.; Kim, H.Y.; Heo, A.J.; Lee, S.H.; Lee, M.J.; Kim, S.B.; Srinivasrao, G.; Mun, S.R.; Cha-Molstad, H.; Ciechanover, A.; et al. The N-Degron Pathway Mediates ER-phagy. Mol Cell 2019, 75, 1058-1072 e1059, doi:10.1016/j.molcel.2019.06.028.
- Zhou, Z.; Liu, J.; Fu, T.; Wu, P.; Peng, C.; Gong, X.; Wang, Y.; Zhang, M.; Li, Y.; Wang, Y.; et al. Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation. Nat Commun 2021, 12, 1570, doi:10.1038/s41467-021-21874-1.
- Grumati, P.; Morozzi, G.; Holper, S.; Mari, M.; Harwardt, M.I.; Yan, R.; Muller, S.; Reggiori, F.; Heilemann, M.; Dikic, I. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. Elife 2017, 6, doi:10.7554/eLife.25555.
- Chino, H.; Hatta, T.; Natsume, T.; Mizushima, N. Intrinsically Disordered Protein TEX264 Mediates ER-phagy. Mol Cell 2019, 74, 909-921 e906, doi:10.1016/j.molcel.2019.03.033.
- Chen, Q.; Xiao, Y.; Chai, P.; Zheng, P.; Teng, J.; Chen, J. ATL3 Is a Tubular ER-Phagy Receptor for GABARAP-Mediated Selective Autophagy. Curr Biol 2019, 29, 846-855 e846, doi:10.1016/j.cub.2019.01.041.
- Fumagalli, F.; Noack, J.; Bergmann, T.J.; Cebollero, E.; Pisoni, G.B.; Fasana, E.; Fregno, I.; Galli, C.; Loi, M.; Solda, T.; et al. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat Cell Biol 2016, 18, 1173-1184, doi:10.1038/ncb3423.
- Nthiga, T.M.; Kumar Shrestha, B.; Sjottem, E.; Bruun, J.A.; Bowitz Larsen, K.; Bhujabal, Z.; Lamark, T.; Johansen, T. CALCOCO1 acts with VAMP-associated proteins to mediate ER-phagy. EMBO J 2020, 39, e103649, doi:10.15252/embj.2019103649.
- Stefely, J.A.; Zhang, Y.; Freiberger, E.C.; Kwiecien, N.W.; Thomas, H.E.; Davis, A.M.; Lowry, N.D.; Vincent, C.E.; Shishkova, E.; Clark, N.A.; et al. Mass spectrometry proteomics reveals a function for mammalian CALCOCO1 in MTOR-regulated selective autophagy. Autophagy 2020, 16, 2219-2237, doi:10.1080/15548627.2020.1719746.
- Rubinsztein, D.C. Receptors for selective recycling. Nature 2015, 522, 291-292, doi:10.1038/nature14532.
- Nakatogawa, H.; Mochida, K. Reticulophagy and nucleophagy: New findings and unsolved issues. Autophagy 2015, 11, 2377-2378, doi:10.1080/15548627.2015.1106665.
- Zhao, D.; Du, L.L. Epr1, a UPR-upregulated soluble autophagy receptor for reticulophagy. Autophagy 2020, 16, 2112-2113, doi:10.1080/15548627.2020.1816665.
- Zhao, D.; Zou, C.X.; Liu, X.M.; Jiang, Z.D.; Yu, Z.Q.; Suo, F.; Du, T.Y.; Dong, M.Q.; He, W.; Du, L.L. A UPR-Induced Soluble ER-Phagy Receptor Acts with VAPs to Confer ER Stress Resistance. Mol Cell 2020, 79, 963-977 e963, doi:10.1016/j.molcel.2020.07.019.
- Stephani, M.; Picchianti, L.; Gajic, A.; Beveridge, R.; Skarwan, E.; Sanchez de Medina Hernandez, V.; Mohseni, A.; Clavel, M.; Zeng, Y.; Naumann, C.; et al. A cross-kingdom conserved ER-phagy receptor maintains endoplasmic reticulum homeostasis during stress. Elife 2020, 9, doi:10.7554/eLife.58396.
- Cui, Y.; Parashar, S.; Zahoor, M.; Needham, P.G.; Mari, M.; Zhu, M.; Chen, S.; Ho, H.C.; Reggiori, F.; Farhan, H.; et al. A COPII subunit acts with an autophagy receptor to target endoplasmic reticulum for degradation. Science 2019, 365, 53-60, doi:10.1126/science.aau9263.
- Cui, Y.; Parashar, S.; Ferro-Novick, S. A new role for a COPII cargo adaptor in autophagy. Autophagy 2020, 16, 376-378, doi:10.1080/15548627.2019.1699347.
- Chen, S.; Novick, P.; Ferro-Novick, S. ER network formation requires a balance of the dynamin-like GTPase Sey1p and the Lunapark family member Lnp1p. Nat Cell Biol 2012, 14, 707-716, doi:10.1038/ncb2523.
- Chen, S.; Cui, Y.; Parashar, S.; Novick, P.J.; Ferro-Novick, S. ER-phagy requires Lnp1, a protein that stabilizes rearrangements of the ER network. Proc Natl Acad Sci U S A 2018, 115, E6237-E6244, doi:10.1073/pnas.1805032115.
- Chen, S.; Mari, M.; Parashar, S.; Liu, D.; Cui, Y.; Reggiori, F.; Novick, P.J.; Ferro-Novick, S. Vps13 is required for the packaging of the ER into autophagosomes during ER-phagy. Proc Natl Acad Sci U S A 2020, 117, 18530-18539, doi:10.1073/pnas.2008923117.
- Mallucci, G.R.; Klenerman, D.; Rubinsztein, D.C. Developing Therapies for Neurodegenerative Disorders: Insights from Protein Aggregation and Cellular Stress Responses. Annu Rev Cell Dev Biol 2020, 36, 165-189, doi:10.1146/annurev-cellbio-040320-120625.




