Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Grazia Licciardello | + 2936 word(s) | 2936 | 2021-09-09 05:31:43 | | | |
| 2 | Rita Xu | -18 word(s) | 2918 | 2021-09-17 05:49:54 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Licciardello, G. Exotic and Emergent Citrus Viruses. Encyclopedia. Available online: https://encyclopedia.pub/entry/14261 (accessed on 08 February 2026).
Licciardello G. Exotic and Emergent Citrus Viruses. Encyclopedia. Available at: https://encyclopedia.pub/entry/14261. Accessed February 08, 2026.
Licciardello, Grazia. "Exotic and Emergent Citrus Viruses" Encyclopedia, https://encyclopedia.pub/entry/14261 (accessed February 08, 2026).
Licciardello, G. (2021, September 16). Exotic and Emergent Citrus Viruses. In Encyclopedia. https://encyclopedia.pub/entry/14261
Licciardello, Grazia. "Exotic and Emergent Citrus Viruses." Encyclopedia. Web. 16 September, 2021.
Copy Citation
Authors focus on exotic and emerging dangerous citrus viruses that have still not been reported in the countries of the Mediterranean area, that are not yet regulated or that are restricted to certain small areas. They also discuss the contribution that old and new technologies may offer for valuable surveys aimed at promoting the adoption and sharing of better control measures and for the production of pathogen-tested citrus trees and rootstocks.
surveillance
detection
high throughput sequencing
EPPO quarantine regulation
diagnosis
1. Introduction
Although there are no cultivated citrus trees with Mediterranean wild-type ancestors, in the last two millennia, citrus trees have become popularly associated with the region, and citrus cultivation continues to have considerable economic and cultural significance throughout the Mediterranean and in surrounding areas. The first Citrus species to reach the East Mediterranean shores was probably the Etrog citron (Citrus medica), brought over from Persia and Medea. Sour oranges (C. aurantium) and lemons (C. lemon) followed, whereas the arrival of sweet oranges (C. sinensis) apparently took place only after 1499, following the discovery of the European–Far East sea-line by Vasco de Gama from Portugal. Indeed, several languages, including Arabic and the Romanian, refer to oranges as ‘portucal’. Interestingly, for decades, the Mediterranean citrus industry flourished with the use of seedling trees, a practice that had to be changed following a further maritime development allowing the transfer of rooted plants and soil infested with Phytophthora. The oomycete that caused this serious decline through disease forced the introduction of grafting on the sour orange rootstocks found by Spanish horticulturists, to end the gummosis epidemic. Similar attempts to use sour orange rootstock conducted in South Africa and the Far East, including Australia and Java (now Indonesia), were unsuccessful due to the wide presence of both citrus tristeza virus (CTV) and its vector. It took years to realize that the use of sour orange as a successful rootstock in handling the gummosis problem was only because citrus plants originally imported from ancient regions were introduced as seeds and, hence, were free from graft-transmitted pathogens.
The considerable growth of citrus demand to supply vitamin C needs on the Europe–Far East routes considerably expanded cultivation, from that in home gardens and orangeries, the luxury structures used until the 19th century to protect orange and other fruit trees during the winter, to more intensive groves. Along with this came an interest in the introduction of new varieties for the botanical gardens that were replacing the private orangeries.
Initially, most of the Mediterranean citrus-producing countries developed their local orange (and, eventually, mandarin) selections. The Jaffa orange, for example, was the most popular variety from about the middle of the nineteenth century, developed in Palestine and later in Israel after its establishment in 1948. This situation continued for at least 30 more years. Other orange varieties, such as the Tarocco and Moro blood oranges, were popular in Sicily and Southern Italy, and other varieties of oranges in Spain and Morocco. A large change occurred with the establishment of the Riverside experiment station in California, a school where many advanced horticulturists throughout the Mediterranean were educated and thus brought home practices of variety collection and variety diversification. With diversification came the import of citrus budwoods infected by different viruses. In many of the citrus areas in the region, the arrival of tristeza occurred 20–30 years before the disease turned into an observable problem. This was partly due to the absence in the region of the most effective, citrus-specific and abundant vector of the virus, Aphis (Toxoptera) citricidus Kirkaldy.
Because, in the early period, the spread of Citrus spp. occurred mainly through fruits and seeds, most of the important citrus disease agents commonly found in China, India and other Far East countries were not carried along. This helped in preventing the spread in the Mediterranean basin of phloem-limited pathogens, such as CTV, and other graft-transmissible viruses and bacteria [1]. Later, in 1920s and 1930s, the expansion of citrus cultivation in the Mediterranean area was associated with the introduction of budwood of exotic citrus varieties. It was later discovered that Meyer lemon imported from China and kumquats and satsumas from Japan led to the geographic dispersion of the tristeza virus, although, at that stage, the disease remained unnoticed there [2]. Worldwide alarming reports were followed with national programs of CTV elimination, which revealed cases of diseased trees in several Mediterranean countries, although still restricted to the original imported propagations. Later, bioindexing and ELISA assays have shown the exchange of propagative material infected by CTV continued for decades [3]. New technologies are helping now to get information about the introduction of other citrus viruses and viroids.
Once the scale of the threat had been perceived, the European Plant Protection Organization (EPPO) decided to conduct a survey of viral and viral-like diseases of citrus plants in the Mediterranean, conducted by the late Prof. I. Reichert (Agriculture Research Station, Rehovot, Israel). The study mission showed that the danger of citrus viral diseases threatened the entire region [4] and highlighted the need to establish uniform methods of indexing, diagnosis and nomenclature. These observations continue to be relevant, especially since globalization resulted in movement of people and goods and considerable advances in novel diagnostic technologies.
The most serious outbreak of tristeza in the Mediterranean region was noticed in the 1960s in Spain, where several millions of trees grafted on sour orange had succumbed to the disease. Replanting the old clone citrus trees with virus-free planting material generated considerable benefits for the citrus industry in terms of quantity and quality of fruits [5]. A few years later, when the epidemic developed in Israel, the relevant organization set up to eradicate the disease, eventually realized that the majority of the CTV isolates were poorly aggressive. However, once CTV became an observed threat, many of the Mediterranean countries established regulatory policies against the continuation of the use of sour orange as a rootstock, in favor of a replacement plan that has, to date, proven to be protective against CTV.
Today, the increased number of viruses and viroids affecting citrus [6], favored by climatic changes and increased activities through global markets, have increased the risk of the introduction of exotic pests and pathogens, which could spread from a single Mediterranean site to the whole region—a region producing more than 21% of the world’s citrus. Nevertheless, the current knowledge of the occurrence and geographical distribution, biological characteristics and molecular biological features of exotic and emergent citrus viruses and viroids relevant to the area is discontinuous and not homogeneous (Figure 1).
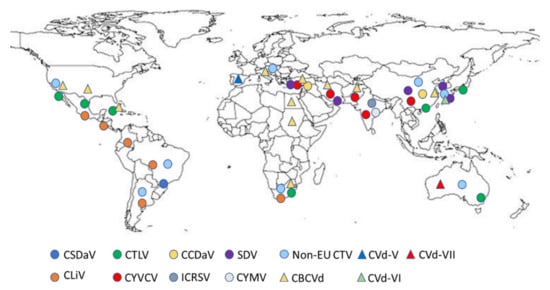
Figure 1. Global distribution map of exotic citrus viruses and viroids relevant to the Mediterranean region (extracted from EPPO Global database 2021, accessed in July 2021). CSDaV, Citrus sudden death-associated virus; CTLV, Citrus tatter leaf virus; CCDaV, Citrus chlorotic dwarf-associated virus; SDV, Satsuma dwarfing virus; Non-EU CTV, stem pitting and resistance breaking citrus tristeza virus; CLiV, Citrus leprosis virus (sensu lato); CYVCV, Citrus yellow vein clearing virus; ICRSV, Indian citrus ringspot virus; CYMV, Citrus mosaic virus; CBCVd, Citrus bark cracking viroid; CVd-V, Citrus viroid V; CVd-VI, Citrus viroid VI; CVd-VII, Citrus viroid VII.
The guidelines for surveillance provided by the International Plant Protection Convention [7] suggest that researchers should consider whether a cooperative effort to collect and record data on pest presence or absence via surveys and monitoring or other procedures [8] would be strategically effective as a means of containing the spread of diseases within a country, as well as to prevent transborder movements among countries [9]. Such a program of surveillance requires a wide knowledge of the complex phytosanitary status of the citrus plants within each country and abroad, in order to focus on pests that are not known to be present in a specific area and to monitor the distribution of that specific pest of interest, or to carry out the identification of cases that would trigger further actions, in line with current international standards and a statistically sound and risk-based pest survey approach [10].
2. Citrus Viruses Recommended for Regulation as Quarantine Pests
2.1. Citrus Tristeza Virus (Closterovirus, Closteroviridae)
Citrus tristeza virus (CTV) is a positive-sense single-stranded genomic RNA of 19.3 kb encapsidated in the p25 major coat protein (95%) and the p27 minor coat protein (5%). The genome codes for 12 open reading frames (ORFs). Its phloem-associated virions consist of filamentous particles about 2000 nm long and 10–12 nm in diameter [11][12].
The host range of CTV is generally limited to Citrus species and their relatives, which are symptomless for many CTV isolates if the virus is on their own roots, although when grafted, they show a wide range of symptoms, depending on the scion–rootstock combination and the virus strains and isolates. The most characterized symptoms are quick decline (QD) and stem pitting (SP). Quick decline, known as tristeza, leads to the death of the plant grafted on sour orange; stem pitting is associated with sparse foliage and reduced yield and fruit quality, regardless of the rootstock. Some rootstocks themselves can be severely affected on the trunk and at the root level (such as alemow). A third symptom, termed seedling yellows (SY), is seen on sour orange, lemon and grapefruit seedlings in the greenhouse or on infected trees regrafted with these varieties [1][5].
Almost all Citrus and Fortunella species host the virus by means of natural infection, and several Rutaceae species, such as Aegle, Aeglopsis, Citropsis and others, as well as non-citrus species (Passiflora gracilis and P. coerulea), have been reported as experimental hosts (see Moreno et al. [5] for comprehensive reference lists) (Table 1). Their roles in the epidemiology of the disease have not been clarified, and this has led to several uncertainties regarding the regulation of their movement as ornamental species [13]. Studies on CTV revealed that the outcome of a CTV infection differs depending on the citrus host and CTV strain [14][15]. In addition, the replication of the virus isolates differs according to the citrus species and variety [16].
Due to the continuous advancement enabled by bioinformatics and new genomic technologies, the possible recombination sites of the CTV 5′ half have been identified, whereas the 3′ half (genes p23, p25, p18, etc.) sites of the genome seem to be far more conserved, probably reflecting the strictness imposed by the functional role of their expression products [14][17]. Distinct members of the same lineage sharing common ancestries share a strain or genotype [14]. The introduction of high-throughput sequencing technologies (HTS) for CTV isolate characterization increased the number of whole genomes sequenced, allowing novel reassessments of the phylogenetic relations of the CTV strains, and the proposal of adding four new strains (S1, L1, M1 and A18) [18][19] to the previous CTV strain catalogue (T36, VT, T3, RB, T68, T30 and HA16-5). To date (as of July 2021), a total of 81 whole-genome sequences of CTV isolates coming from different countries have been listed in the GenBank database [14][15][19][20][21][22][23][24][25][26][27][28][29][30][31][32][33][34][35][36][37][38][39][40][41]. Most of them are representative of the VT and RB genotypes followed by T30 and T36 (Figure 2). Only 13 isolates from the Mediterranean area have been fully sequenced. Nine of them are VT genotypes, two are T30, and two are T36. These figures indicate that our knowledge of the genetic structure of the CTV population in the Mediterranean region is limited compared with that of the wider global populations and is probably incomplete, with potential considerable epidemiological phytosanitary implications.
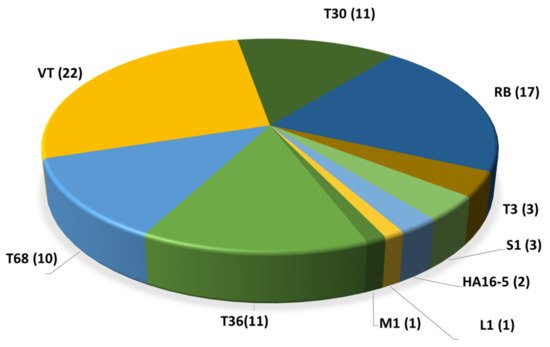
Figure 2. Distribution of the 81 full genome sequences of CTV per genotype available in the GenBank (as of July 2021).
Regarding the epidemiology of the disease in the Mediterranean region, the virus is efficiently vectored by the cotton/melon aphid Aphis gossypii, whereas the more abundant citrus-infesting aphids A. spiraecola (formerly A. citricola) and Aphis (Toxoptera) aurantii in the region seem far less efficient under experimental conditions ([5][42][43][44][45][46][47][48][49][50]). A. citricidus, the most efficient vector of CTV worldwide [5], apparently remains restricted to the island of Madeira and the coastal area of the northwest quadrant of the Iberian Peninsula of Spain and Portugal. Aphids transmit the virus in a semi-persistent mode. The efficiency of transmission also varies between CTV isolates; isolates ST and VT-1, both of which belong to the VT strain, differ considerably in their rates of transmission by A. gossypii [51][52].
The failure of CTV eradication efforts throughout most of the Mediterranean countries, mainly due to the complex patterns of citriculture, in terms of ownership, maintenance and variety composition, has prompted the adaption of novel horticultural practices based on CTV-tolerant rootstocks, such as Troyer and Carrizo citrange, and this is in progress in many countries. Considerable difficulties were noticed, however, due to the underestimation of the new disease, and agronomic problems linked to the soil and water type and/or the fact that some budwood was infected by viroids.
This effective replacement strategy assumes that SP- and RB-inducing isolates and/or their efficient vectors are still absent from the Mediterranean area. Indeed a regional surveillance program should be adopted by the national phytosanitary services to prevent their introduction and spread.
The case of tristeza stem pitting. A CTV disease named stem pitting (SP) has been reported in many countries in the southern hemisphere, where it is responsible for considerable damage. The affected trees of sweet orange, grapefruit and lime species show stunting, low yield and poor fruit quality, regardless of the rootstock used (Figure 3). SP is induced by most CTV isolates when inoculated on Mexican lime and C. macrophylla, indicator plants or rootstocks. Grapefruit, sweet orange and other citrus species may also show this symptom in the field when infected by VT stem pitting isolates. Countries in which the SP isolates are prevalent, causing tree decline and poor performance in crops—as in the case of grapefruits in South Africa and the Pera orange variety in Brazil—have adopted mild isolate cross protection as a practical means of preventing the damaging effects of locally spreading native severe stem-pitting CTV isolates [53][54].

Figure 3. Symptoms of tristeza stem pitting caused by SP isolates of CTV on the trunk (a), stem (b) and branch (c) of grapefruit. Similar effects may occur on some sweet orange varieties and on indicator plants, or on alemow used as rootstock (d).
However, field symptoms of stempitting disease in grapefruit trees have been only occasionally detected in the Eu-Med region, although they have been detected in Mexican lime and alemow seedlings and grapefruit and acidless pummelo plants in greenhouse tests [33][36].
Biological indexing on sweet orange and grapefruit in greenhouse allows to discriminate SP isolates and to evaluate their severity [31].
Resistance-breaking (RB) CTV isolates. Old surveys led researchers to assume that P. trifoliata plants were tolerant or resistant to most of the CTV isolates and strains, due to specific loci that are able to prevent the early steps of the infection process and to modify its expression. A similar mechanism has been reported for Swinglaea glutinosa and Severinia buxifolia. In recent years, certain CTV genotypes, known as resistance-breaking (RB) genotypes, were phylogenetically analyzed and found to belong to at least two subgroups, which shared 90.3% genomic nucleotide sequence identity with the T36 clade [40].
After a T36 isolate of CTV was able to infect and replicate in P. trifoliata protoplasts [55] and the T68 and VT genotypes were detected in the root of P. trifoliata cv. ‘Rubidoux’, the low detection rate for the CTV RB isolates was related to a low titer of the accumulated virus or an uneven distribution in shoots [40]. This limitation was apparently surmounted when a VT strain was found to coinfect a tree, generating a positive interaction for the infection [56][57].
After the discovery of a CTV isolate capable of breaking CTV resistance in trifoliate oranges and replicating in roots, bark and foliage, the spread of these isolates was investigated in many countries outside the Mediterranean region [13], and they were recently found to have spread in China [58]. A few infected trees have been found in Morocco and rogued [59]. The productivity of infected trees is variable in relation to the scion–rootstock combination and the mixture of CTV isolates infecting the tree.
Different molecular methods for the detection of RB isolates are available. The first identification method using RT-PCR was described in 2013 by Roy et al. [39], who developed a genotype-specific assay that enabled them to overcome the difficulties of the MMM method to detect the presence of an unassigned genotype such as RB. However, in 2015, a new MMM marker specific for RB, targeting the ORF1a K17 region, was designed and used to study the diversity of the CTV strains present in New Zealand and the Pacific [56]. Cook et al. [60] proposed an RT-PCR protocol with three pairs of primers specifically designed to amplify RB group 1, RB group 2 and HA16-5, targeting a portion of the LProII domain of ORF1a. This protocol has been successfully used to detect multiple strains in two South African cross-protecting sources. A lab-on-chip miniaturized platform based on a sequential multiplex RT-PCR and microarray hybridization allowed researchers to simultaneously discriminate between the T36, T30, VT, RB, T68 and T3 strains in mixed infections [61].
Saponari et al. [62] describe a protocol developed in California, based on a preliminary assay related to the Mab MCA13 and T36NS probe, designed in the intergenic region between p25 and p27, followed by a bark inoculation on P. trifoliata and a test on Madam Vinous seedlings after a passage on trifoliate orange to check the infectivity, and a p65 sequence phylogenetic analysis.The full-length genome sequencing of these isolates confirmed the phylogenetic analysis. Two subclades were distinguished, with Californian isolates falling in both, and the New Zealand isolates placed together in group I, differently from the findings of Cook et al. [60].
References
- Bar-Joseph, M.; Batuman, O.; Roistacher, N. The history of citrus tristeza virus–Revisited. In Citrus Tristeza Virus Complex and Tristeza Diseases; Karasev, A.V., Hilf, M., Eds.; APS Press: Saint Paul, MN, USA, 2009; pp. 3–26.
- Bar-Joseph, M.; Catara, A. Endemic and emerging vector-borne Mediterranean citrus diseases and their epidemiological consequences. In Integrated Control of Citrus Pests in the Mediterranean Region; Vacante, V., Gerson, V., Eds.; Bentham Science Pubblishers Ltd.: Sharjah, UAE, 2010; pp. 137–155.
- Davino, S.; Willemsen, A.; Panno, S.; Davino, M.; Catara, A.; Elena, S.F.; Rubio, L. Emergence and phylodynamics of Citrus tristeza virus in Sicily, Italy. PLoS ONE 2013, 8, e66700.
- Reichert, I. EPPO study mission to investigate virus diseases of citrus in the Mediterranean countries. In Report of the International Conference on Virus Diseases of Citrus (Acireale, Sicily, 15–17 September 1959); European and Mediterranean Plant Protection Organisation: Paris, France, 1960; pp. 13–44.
- Moreno, P.; Ambros, S.; Albiach-Martì, M.R.; Guerri, J.; Peña, L. Citrus tristeza virus: A pathogen that changed the course of the citrus industry. Mol. Plant Path. 2008, 9, 251–268.
- Zhou, C.; da Graça, J.V.; Freitas-Astúa, J.; Vidalakis, G.; Duran-Vila, N.; Lavagi, I. Citrus viruses and viroids. In The Genus Citrus, 2nd ed.; Talon, M., Caruso, M., Gmitter, F., Jr., Eds.; Woodhead Publishing: Cambridge, UK, 2020; pp. 391–410.
- FAO (Food and Agriculture Organization of the United Nations). Plant Pest Surveillance: A Guide to Understand the Principal Requirements of Surveillance Programmes for National Plant Protection Organizations; FAO: Rome, Italy, 2016.
- ISPM 5. Glossary of Phytosanitary Terms; IPPC/FAO: Rome, Italy, 2017.
- ISPM 6. Surveillance; IPPC/FAO: Rome, Italy, 2018.
- EFSA (European Food Safety Authority); Schrader, G.; Camilleri, M.; Diakaki, M.; Vos., S. Pest survey card on non-European isolates of citrus tristeza virus. EFSA Support. Publ. 2019, 16, 1600E.
- Kitajima, E.W.; Silva, D.M.; Oliveira, A.R.; Müller, G.W.; Costa, A.S. Thread-like particles associated with tristeza disease of citrus. Nature 1964, 201, 1011–1012.
- Martelli, G.P.; Agranosky, A.A.; Bar Joseph, M.; Boscia, D.; Candresse, T.; Coutts, R.H.A.; Dolja, V.V.; Hu, J.S.; Jelkmann, W.; Karasev, A.V.; et al. Family Closteroviridae. In Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier-Academic Press: Amsterdam, The Netherlands, 2011; pp. 987–1001.
- EFSA PLH Panel (EFSA Panel on Plant Health). Scientific opinion on the pest categorisation of Citrus tristeza virus (non-European isolates). EFSA J. 2017, 15, 5031.
- Harper, S.J. Citrus tristeza virus: Evolution of complex and varied genotypic groups. Front. Microbiol. 2013, 4, 93.
- Yokomi, R.K.; Selvaraj, V.; Maheshwari, Y.; Saponari, M.; Giampetruzzi, A.; Chiumenti, M.; Hajeri, S. Identification and characterization of CTV RB strains Citrus tristeza virus isolates breaking resistance in trifoliate orange in California. Phytopathology 2017, 107, 901–908.
- Targon, M.L.P.N.; Machado, M.A.; Muller, G.W.; Coletta Filho, H.D.; Manjunath, K.L.; Lee, R.F. Sequence of coat protein gene of the severe Citrus tristeza virus complex Capão Bonito. In Proceedings of the 14th Conference of International Organization of Citrus Virologists (IOCV), Campinas, Brazil, 13–18 September 1998; da Graca, J.V., Lee, R.F., Yokomi, R.K., Eds.; IOCV: Riverside, CA, USA, 2000; pp. 121–126.
- Roy, A.; Ananthakrishnan, G.; Hartung, J.S.; Brlansky, R.H. Development and application of a multiplex reverse transcription polymerase chain reaction assay for screening a global collection of Citrus tristeza virus isolates. Phytopathology 2010, 100, 1077–1088.
- Bester, R.; Cook, G.; Maree, H.J. Citrus tristeza virus genotype detection using high-throughput sequencing. Viruses 2021, 13, 168.
- Yokomi, R.; Selvaraj, V.; Maheshwari, Y.; Chiumenti, M.; Saponari, M.; Giampetruzzi, A.; Weng, Z.; Xiong, Z.; Hajeri, S. Molecular and biological characterization of a novel mild strain of citrus tristeza virus in California. Arch. Virol. 2018, 163, 1795–1804.
- Karasev, A.V.; Boyko, V.P.; Gowda, S.; Nikolaeva, O.V.; Hilf, M.E.; Koonin, E.V.; Niblett, C.L.; Cline, K.; Gumpf, D.J.; Lee, R.F.; et al. Complete sequence of Citrus tristeza virus RNA genome. Virology 1995, 208, 511–520.
- Chen, A.Y.S.; Watanabe, S.; Yokomi, R.; Ng, J.C.K. Nucleotide heterogeneity at the terminal ends of the genomes of two California Citrus tristeza virus strains and their complete genome sequence analysis. Virol. J. 2018, 15, 141.
- Pappu, H.R.; Karasev, A.V.; Anderson, E.J.; Pappu, S.S.; Hilf, M.E.; Febres, V.J.; Eckloff, R.M.; McCaffery, M.; Boyko, V.; Gowda, S.; et al. Nucleotide sequence and organization of eight 3’ open reading frames of the citrus tristeza closterovirus genome. Virology 1994, 199, 35–46.
- Scuderi, G.; Russo, M.; Davino, S.; Ferraro, R.; Catara, A.; Licciardello, G. Occurrence of the T36 Genotype of Citrus tristeza virus in Citrus Orchards in Sicily, Italy. Plant Dis. 2016, 100, 1253.
- Weng, Z.; Barthelson, R.; Gowda, S.; Hilf, M.E.; Dawson, W.O.; Galbraith, D.W.; Xiong, Z. Persistent infection and promiscuous recombination of multiple genotypes of an RNA virus within a single host generate extensive diversity. PLoS ONE 2007, 2, e917.
- Roy, A.; Brlansky, R.H. Genome analysis of an orange stem pitting Citrus tristeza virus isolate reveals a novel recombinant genotype. Virus Res. 2010, 151, 118–130.
- Cook, G.; Breytenbach, J.H.J.; Steyn, C.; de Bruyn, R.; van Vuuren, S.P.; Burger, J.T.; Maree, H.J. Grapefruit field trial evaluation of Citrus tristeza virus T68-strain sources. Plant Dis. 2021, 105, 361–367.
- Harper, S.J.; Dawson, T.E.; Pearson, M.N. Complete genome sequences of two distinct and diverse Citrus tristeza virus isolates from New Zealand. Arch. Virol. 2009, 154, 1505–1510.
- Zablocki, O.; Pietersen, G. Characterization of a novel citrus tristeza virus genotype within three cross-protecting source GFMS12 sub-isolates in South Africa by means of Illumina sequencing. Arch. Virol. 2014, 159, 2133–2139.
- Licciardello, G.; Scuderi, G.; Ferraro, R.; Giampetruzzi, A.; Russo, M.; Lombardo, A.; Raspagliesi, D.; Bar-Joseph, M.; Catara, A. Deep sequencing and analysis of small RNAs in sweet orange grafted on sour orange infected with two Citrus tristeza virus isolates prevalent in Sicily. Arch. Virol. 2015, 160, 2583–2589.
- Mawassi, M.; Mietkiewska, E.; Gofman, R.; Yang, G.; Bar-Joseph, M. Unusual sequence relationships between two isolates of Citrus tristeza virus. J. Gen. Virol. 1996, 77, 2359–2364.
- Licciardello, G.; Ferraro, R.; Scuderi, G.; Russo, M.; Catara, A.F. A simulation of the use of high throughput sequencing as pre-screening assay to enhance the surveillance of citrus viruses and viroids in the EPPO region. Agriculture 2021, 11, 400.
- Varveri, C.; Olmos, A.; Pina, J.A.; Marroquín, C.; Cambra, M. Biological and molecular characterization of a distinct Citrus tristeza virus isolate originating from a lemon tree in Greece. Plant Pathol. 2015, 64, 792–798.
- Ruiz-Ruiz, S.; Moreno, P.; Guerri, J.; Ambrós, S. The complete nucleotide sequence of a severe stem pitting isolate of Citrus tristeza virus from Spain: Comparison with isolates from different origins. Arch. Virol. 2006, 151, 387–398.
- Biswas, K.K.; Tarafdar, A.; Sharma, S.K. Complete genome sequence of mandarin decline Citrus tristeza virus of the north-Eastern Himalayan hill region of India: Comparative analyses determine recombinant. Arch. Virol. 2012, 157, 579–583.
- Matsumura, E.E.; Coletta-Filho, H.D.; Nouri, S.; Falk, B.W.; Nerva, L.; Oliveira, T.S.; Dorta, S.O.; Machado, M.A. Deep sequencing analysis of RNAs from citrus plants grown in a citrus sudden death-affected area reveals diverse known and putative novel viruses. Viruses 2017, 9, 92.
- Yang, Z.N.; Mathews, D.M.; Dodds, J.A.; Mirkov, T.E. Molecular characterization of an isolate of citrus tristeza virus that causes severe symptoms in sweet orange. Virus Genes 1999, 19, 131–142.
- Albiach-Marti, M.R.; Mawassi, M.; Gowda, S.; Satanarayana, T.; Hilf, M.E.; Shanker, S.; Almira, E.C.; Vives, M.C.; López, C.; Guerri, J.; et al. Sequences of Citrus tristeza virus separated in time and space are essentially identical. J. Virol. 2000, 74, 6856–6865.
- Vives, M.C.; Rubio, L.L.; Pez, C.; Navas-Castillo, J.; Albiach-Mart, M.R.; Dawson, W.O.; Guerri, J.; Flores, R.; Moreno, P. The complete genome sequence of the major component of a mild citrus tristeza virus isolate. J. Gen. Virol. 1999, 80, 811–816.
- Roy, A.; Choudhary, N.; Hartung, J.S.; Brlansky, R.H. The prevalence of the Citrus tristeza virus trifoliate resistance breaking genotype among Puerto Rican isolates. Plant Dis. 2013, 97, 1227–1234.
- Harper, S.J.; Dawson, T.E.; Pearson, M.N. Isolates of Citrus tristeza virus that overcome Poncirus trifoliata resistance comprise a novel strain. Arch. Virol. 2010, 155, 471–480.
- Melzer, M.J.; Borth, W.B.; Sether, D.M.; Ferreira, S.; Gonsalves, D.; Hu, J.S. Genetic diversity and evidence for recent modular recombination in Hawaiian Citrus tristeza virus. Virus Genes 2010, 40, 111–118.
- Maccheroni, W.; Alegria, M.C.; Greggio, C.C.; Piazza, J.P.; Kamla, R.F.; Zacharias, P.R.A.; Bar-Joseph, M.; Kitajima, E.W.; Assumpção, L.C.; Camarotte, G.; et al. Identification and genomic characterization of a new virus (Tymoviridae family) associated with citrus sudden death disease. J. Virol. 2005, 79, 3028–3037.
- Afloukou, F.M.; Çalişkan, F.; Önelge, N. Aphis gossypii Glover is a vector of citrus yellow vein clearing virus. J. Gen. Plant Pathol. 2021, 87, 83–86.
- Onelge, N.; Satar, S.; Elibüyük, O.; Bozan, O.; Kamberoglu, M. Transmission studies on Citrus yellow vein clearing virus. In Proceedings of the 18th Conference International Organization of Citrus Virology, Campinas, Brazil, 7–12 November 2010; da Graca, J.V., Lee, R.F., Yokomi, R.K., Eds.; IOCV: Riverside, CA, USA, 2011.
- Zhang, Y.H.; Wang, Y.L.; Qin, W.; Cao, M.J.; Zhou, C.Y.; Yan, Z. Identification of Aphis spiraecola as a vector of Citrus yellow vein clearing virus. Eur. J. Plant Pathol. 2018, 152, 841–844.
- Bastianel, M.; Novelli, V.M.; Kitajima, E.W.; Kubo, K.S.; Bassanezi, R.B.; Machado, M.A.; Freitas-Astua, J. Citrus Leprosis: Centennial of an unusual mite-virus pathosystem. Plant Dis. 2010, 94, 284–292.
- Kubo, K.S.; Novelli, V.M.; Bastianel, M.; Locali-Fabris, E.C.; Antonioli-Luizon, R.; Machado, M.A.; Freitas-Astua, J. Detection of Brevipalpus-transmitted viruses in their mite vectors by RT–PCR. Exp. Appl. Acarol. 2011, 54, 33–39.
- Zhang, Y.H.; Liu, C.H.; Wang, Q.; Wang, Y.L.; Zhou, C.Y.; Zhou, Y. Identification of Dialeurodes citri as a vector of citrus yellow vein clearing virus in China. Plant Dis. 2019, 103, 65–68.
- Korkmaz, S.; Çinar, A.; Kersting, U.; Garnsey, S.M. Citrus chlorotic dwarf: A new whitefly-transmitted virus-like disease of citrus in Turkey. Plant Dis. 1995, 79, 1074.
- Garnsey, S.M.; Behe, C.G.; Lockhart, B.E.L. Transmission of citrus yellow mosaic badnavirus by the citrus mealybug. Phytopathology 1998, 9, S43.
- Raccah, B.; Loebenstein, G.; Bar-Joseph, M. Transmission of citrus tristeza virus by melon aphid. Phytopathology 1976, 66, 1102–1104.
- Bar-Joseph, M.; Loebenstein, G. Effects of strain, source plant, and temperature on the transmissibility of citrus tristeza virus by the melon aphid. Phytopathology 1973, 63, 716–720.
- Costa, A.S.; Muller, G.W. Tristeza control by cross protection: A U.S.-Brasil cooperative success. Plant Dis. 1980, 64, 538–541.
- Cook, G.; van Vuuren, S.P.; Breytenbach, J.H.J.; Steyn, C.; Burger, J.T.; Maree, H.J. Characterization of Citrus tristeza virus single-variant sources in grapefruit in greenhouse and field trials. Plant Dis. 2016, 100, 2251–2256.
- Albiach-Marti, M.R.; Grosser, J.W.; Gowda, S.; Mawassi, M.; Satyanarayana, T.; Garnsey, S.M.; Dawson, W.O. Citrus tristeza virus replicates and forms infections virions in protoplasts of resistant citrus relatives. Mol. Breed. 2004, 14, 117–128.
- Harper, S.J.; Pearson, M.N. Citrus tristeza virus strains present in New Zealand and the South Pacific. J. Cit. Pathol. 2015, 2, 1–5.
- Zhou, Y.; Liu, Y.; Liu, K.; Yang, F.; Zhou, C. Distribution and population structure of Citrus tristeza virus in Poncirus trifoliata. Austral. Plant Pathol. 2017, 46, 351–355.
- Wang, J.; Zhou, T.; Cao, M.; Zhou, Y.; Li, Z. First report of citrus tristeza virus trifoliate resistance-breaking (RB) genotype in Citrus grandis in China. J. Plant Pathol. 2019, 101, 451.
- Afechtal, M.; D’Onghia, A.M.; Cocuzza, G.E.M.; Djelouah, K. First report of the Citrus tristeza virus resistance-breaking strain in Morocco. J. Plant Path. 2018, 100, 351.
- Cook, G.; van Vuuren, S.P.; Breytenbach, J.H.J.; Burger, J.T.; Maree, H.J. Expanded strain-specific RT-PCR assay for differential detection of currently known Citrus tristeza virus strains: A useful screening tool. J. Phytopathol. 2016, 164, 847–851.
- Scuderi, G.; Catara, A.F.; Licciardello, G. Genotyping citrus tristeza virus isolates by sequential multiplex RT-PCR and microarray hybridization in a lab-on-chip device. In Citrus Tristeza Virus: Methods in Molecular Biology; Catara, A., Bar-Joseph, M., Licciardello, G., Eds.; Humana: New York, NY, USA, 2019; Volume 2015, pp. 127–142.
- Saponari, M.; Giampetruzzi, A.; Selvaraj, V.; Maheshwari, Y.; Yokomi, R. Identification and characterization of resistance-breaking (RB) isolates of citrus tristeza virus. In Citrus Tristeza Virus: Methods in Molecular Biology; Catara, A., Bar-Joseph, M., Licciardello, G., Eds.; Humana: New York, NY, USA, 2019; Volume 2015, pp. 105–126.
More
Information
Subjects:
Agriculture, Dairy & Animal Science
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
781
Revisions:
2 times
(View History)
Update Date:
17 Sep 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




