Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Wang, P. Sensing Tyrosinase Activity. Encyclopedia. Available online: https://encyclopedia.pub/entry/13985 (accessed on 08 February 2026).
Wang P. Sensing Tyrosinase Activity. Encyclopedia. Available at: https://encyclopedia.pub/entry/13985. Accessed February 08, 2026.
Wang, Ping. "Sensing Tyrosinase Activity" Encyclopedia, https://encyclopedia.pub/entry/13985 (accessed February 08, 2026).
Wang, P. (2021, September 08). Sensing Tyrosinase Activity. In Encyclopedia. https://encyclopedia.pub/entry/13985
Wang, Ping. "Sensing Tyrosinase Activity." Encyclopedia. Web. 08 September, 2021.
Copy Citation
Tyrosinase (TYR, E.C. 1.14.18.1), a critical enzyme participating in melanogenesis, catalyzes the first two steps in melanin biosynthesis including the ortho-hydroxylation of L-tyrosine and the oxidation of L-DOPA. Previous pharmacological investigations have revealed that an abnormal level of TYR is tightly associated with various dermatoses, including albinism, age spots, and malignant melanoma.
tyrosinase (TYR)
enzymatic activity
optical substrates
TYR inhibitors
high-throughput screening
1. Introduction
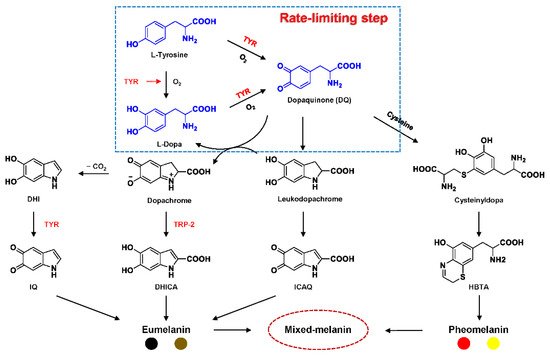
Tyrosinase (TYR, E.C. 1.14.18.1), a type-3 binuclear copper-containing oxidoreductase, efficiently catalyzes o-hydroxylation of monophenols to diphenols (monophenolase activity) and the oxidation of diphenols to quinones (diphenolase activity), without any additional cofactors (Figure 1) [1][2]. It is ubiquitously distributed in organisms ranging from bacteria to eukaryotes and plays a pivotal role in the enzymatic browning of fruit or fungi, as well as mammalian melanin synthesis [3][4]. In mammals, melanin is exclusively synthesized in melanosomes via complex biochemical reactions (Figure 2), and this endogenous substance is primarily responsible for the pigmentation of retina and skin [5][6]. TYR catalyzes the first two steps in melanin biosynthesis: the o-hydroxylation of L-tyrosine and the oxidation of L-DOPA. Since the remainder of the reaction sequence can proceed spontaneously at physiological pH, the conversion of L-tyrosine to dopaquinone (DQ) has been implicated as a crucial rate-limiting procedure in melanogenesis [7][8]. DQ could spontaneously convert into dopachrome, which gradually decomposes into 5,6-dihydroxyindole (DHI) and 5,6-dihydroxyindole-2-carboxylic acid (DHICA) through a succession of redox reactions [5][9]. Ultimately, these dihydroxyindoles are oxidized to eumelanin. Alongside, in the presence of cysteine or glutathione, DQ is converted to 5-S-cysteinyl dopa or glutothionyl dopa, finally yielding pheomelanin [10][11]. The types and relative amounts of these melanin constitute color-based ethnic diversification. Three tyrosinase-like enzymes co-regulate melanogenesis, including TYR and TYR-related proteins 1 (TRP-1) and 2 (TRP-2). TRP-1 shows DHICA oxidase and low tyrosine hydroxylase activity when zinc is replaced by copper. TRP-2 contains two zinc ions at the active site and isomerizes dopachrome to DHICA. They are metal-containing glycoproteins and share ~40% amino acid sequence identity and ~70% similarity [1]. Despite TYR and TYR-related proteins 1 (TRP-1) and 2 (TRP-2) being necessary for melanogenesis, TYR is the most critical rate-limiting enzyme [10].
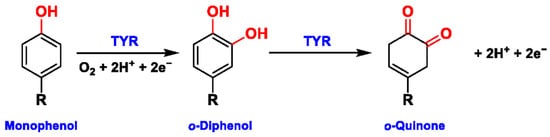
Figure 1. The slow ortho-hydroxylation of monophenol and the fast oxidation of catechol catalyzed by TYR.
Recent pharmacological investigations have revealed that the abnormal metabolism of melanocytes and imbalance in TYR activity are indirectly or directly responsible for various dermatoses [1][12][13]. For example, functional mutations in the gene encoding TYR (TYR, 11q14-21, MIM 606933) would inactivate TYR and cause a deficiency in melanin, thereby resulting in oculocutaneous albinism type 1 (OCA1, MIM 203100), an autosomal recessive disorder characterized by the absence of pigment in hair, skin, and eyes [14]. On the contrary, excess melanin accumulation or abnormal distribution would give rise to hyperpigmentation disorders, including age spots, post-inflammatory hyperpigmentation, and even malignant melanoma [13][15]. In particular, the overexpression of TYR and TRP 1 is significantly associated with the risk of melanoma, a fatal skin carcinoma [16][17][18]. To this end, it has been viewed as a relatively specific biomarker and therapeutic target for melanoma lesions. Additionally, the abnormal level of TYR induces dopamine neurotoxicity and neurodegeneration, which is related to Parkinson’s disease (PD) [19][20][21]. Monitoring TYR activity in complex biosystems undoubtedly remains critical and challenging for biomedical research and drug high-throughput screening (HTS).
In recent decades, various analytical techniques, including immunochemical analysis, mass spectrometry-based proteomics, and substrate-based biochemical assays, have flourished for the quantification of TYR. However, only the substrate-based biochemical assay could rapidly and sensitively determine the real activity of TYR in complex biological systems, leading to its common use in drug discovery and clinical studies [22].
2. Biochemical Characteristics of TYR
2.1. Structural Feature and Catalytic Mechanism of TYR
Human TYR is a glycoprotein (13% carbohydrate) predominantly located in the melanosome membrane of melanocytes [23][24]. The presence of the transmembrane domain and glycans renders it difficult to isolate homogeneous TYR from melanocytes, which impedes crystallographic studies. Fortunately, the most characteristic TYR can be acquired from Streptomyces glausescens, the fungi Neurospore crassa, or Agaricus bisporus [7][25][26]. To compare the conservation of catalytic cavity, the crystal structures of TYR from bacteria (Streptomyces castaneoglobisporus [25] and Bacillus megaterium [27]), fungi Agaricus bisporus [28], and walnut leaves [29] were retrieved from Protein Date Bank. A salient feature of TYR from various sources (Figure 3) is the presence of strictly conserved binuclear copper atoms at the active site, each of copper atoms is coordinated with three conserved histidines, respectively [3]. Moreover, the normal redox state of copper atoms is exceedingly significant for enzyme activity. Due to the relatively high similarity and homology with mammalian TYR, mushroom TYR from Agaricus bisporus acts as a model for enzyme kinetics and inhibitor screening [28][30][31]. Although extensive studies are devoted to TYR, its catalytic mechanism remains controversial. According to the presence/absence of oxygen and the oxidation state of copper ions [Cu (Ⅱ)/Cu (Ⅰ)], three enzymatic forms (Eoxy, Emet, and Edeoxy) participate in the catalytic cycle (Figure 4) [3][32]. The resting form of TYR is found to be a mixture of 85% met and 15% oxy forms, while only the latter could act on the monophenol [7][33][34]. During the monophenolase cycle, to form Eoxy-monophenol complex (EoxyM), the oxygen atom on the deprotonated monophenol is coordinated with the coppers of Eoxy. Then, the phenol is o-hydroxylated to generate Emet-diphenol (D) complex (EmetD) [26][35]. Reducing agents could well draw the Emet into the Edeoxy, with concomitant oxidation to the corresponding o-quinone [36]. Since the deoxy form is the only one capable of reacting with oxygen to regenerate Eoxy and continue the catalytic action, monophenolase activity usually manifests as a characteristic lag time until a sufficient amount of catechol helps Emet to become Edeoxy [36]. Remarkably, this period depends on several factors, including enzyme concentration, monophenol concentration and the presence of reducing agents, especially o-diphenol derivatives (such as L-DOPA) that could shorten and even abolish the lag time [30][33][37]. In the diphenolase cycle, Eoxy continues to bind o-diphenol to originate the EoxyD complex, while both Eoxy and Emet are capable of oxidizing the diphenol to the o-quinone. After this, Emet is regenerated to complete the catalytic cycle continuously [30][34].
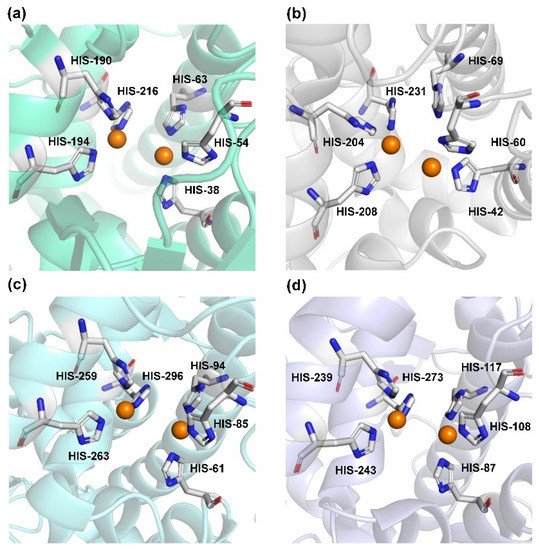
Figure 3. The conserved cavity of TYR from different sources. (a) The crystal of TYR from Streptomyces castaneoglobisporus (PDB ID: 2ZMX). (b) The crystal of TYR from Bacillus megaterium (PDB ID: 3NQ1). (c) The Crystal of TYR from fungus (PDB ID: 2Y9W, Agaricus bisporus). (d) The Crystal of TYR from plant (PDB ID: 5CE9, Juglans regia). Two copper ions (orange) are coordinated with three histidine residues, respectively.
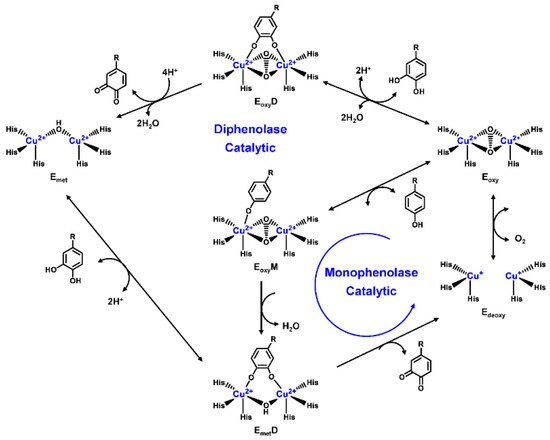
Figure 4. Catalytic cycle of TYR.
2.2. Substrate Specificity of TYR
Based on the broad substrate spectrum, in principle, any simple monophenol or corresponding catechol appears to be its substrate [38]. Besides, TYR also oxidizes various aromatic amines, o-aminophenols, and aromatic o-diamines (Figure 5), despite the reaction rates being orders of magnitude smaller than the corresponding phenols or catechol [39][40]. In terms of phenols, mammalian TYR tends to be relatively specific for its physiological substrate (L-tyrosine and L-DOPA) and has a higher affinity for the L-isomers [41]. A prevalent characteristic in monophenol substrates is without substituents in the ortho-position of the phenolic hydroxyl group. Understandably, large side-chain substituents increase the difficulty of substrate interaction with the key catalytic residues; this is unpropitious for the recognition and catalytic process between the enzyme and ligand [36]. A kinetic study [42] quantitatively discussed the effects of substituents in the 1-position of the aromatic ring on the rate of hydroxylation catalyzed by TYR. The results revealed that monophenols with a high electron donor tend to be oxidized faster [42]. In sharp contrast, the oxidation rate of catechol is positively correlated with the electron-withdrawing capacity of the para-substituents [36]. As such, the steric hindrance, stereochemical characteristics, and electronic effects of substituents have a distinct influence on the rate of TYR-mediated catalysis.
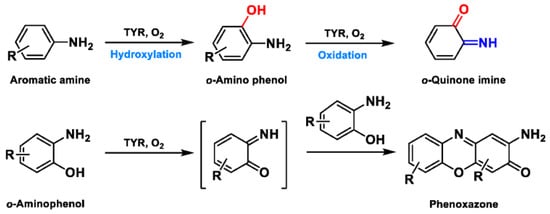
Figure 5. The catalytic reaction of TYR-mediated aromatic amine and o-aminophenol. Adapted with permission from ref. [43]. 1987, American Chemical Society.
References
- Lai, X.; Wichers, H.J.; Soler-Lopez, M.; Dijkstra, B.W. Structure and Function of Human Tyrosinase and Tyrosinase-Related Proteins. Chemistry 2018, 24, 47–55.
- Min, K.; Park, G.W.; Yoo, Y.J.; Lee, J.S. A perspective on the biotechnological applications of the versatile tyrosinase. Bioresour. Technol. 2019, 289, 121730.
- Olivares, C.; Solano, F. New insights into the active site structure and catalytic mechanism of tyrosinase and its related proteins. Pigment. Cell Melanoma Res. 2009, 22, 750–760.
- Lee, S.H.; Baek, K.; Lee, J.E.; Kim, B.G. Using tyrosinase as a monophenol monooxygenase: A combined strategy for effective inhibition of melanin formation. Biotechnol. Bioeng. 2016, 113, 735–743.
- Pillaiyar, T.; Manickam, M.; Jung, S.H. Downregulation of melanogenesis: Drug discovery and therapeutic options. Drug Discov. Today 2017, 22, 282–298.
- Brenner, M.; Hearing, V.J. The protective role of melanin against UV damage in human skin. Photochem. Photobiol. 2008, 84, 539–549.
- Chang, T.S. An updated review of tyrosinase inhibitors. Int. J. Mol. Sci. 2009, 10, 2440–2475.
- Pillaiyar, T.; Namasivayam, V.; Manickam, M.; Jung, S.H. Inhibitors of Melanogenesis: An Updated Review. J. Med. Chem. 2018, 61, 7395–7418.
- Lee, S.Y.; Baek, N.; Nam, T.G. Natural, semisynthetic and synthetic tyrosinase inhibitors. J. Enzyme Inhib. Med. Chem. 2016, 31, 1–13.
- Pillaiyar, T.; Manickam, M.; Namasivayam, V. Skin whitening agents: Medicinal chemistry perspective of tyrosinase inhibitors. J. Enzyme Inhib. Med. Chem. 2017, 32, 403–425.
- Hearing, V.J.; Tsukamoto, K. Enzymatic control of pigmentation in mammals. FASEB J. 1991, 5, 2902–2909.
- Ando, H.; Ichihashi, M.; Hearing, V.J. Role of the ubiquitin proteasome system in regulating skin pigmentation. Int. J. Mol. Sci. 2009, 10, 4428–4434.
- Costin, G.E.; Hearing, V.J. Human skin pigmentation: Melanocytes modulate skin color in response to stress. FASEB J. 2007, 21, 976–994.
- Ray, K.; Chaki, M.; Sengupta, M. Tyrosinase and ocular diseases: Some novel thoughts on the molecular basis of oculocutaneous albinism type 1. Prog. Retin. Eye Res. 2007, 26, 323–358.
- dos Santos, V.I.F.; Lima, M.D.F.; Sofia, M. Mechanisms regulating melanogenesis. An. Bras. Dermatol. 2013, 88, 76–83.
- Ghanem, G.; Fabrice, J. Tyrosinase related protein 1 (TYRP1/gp75) in human cutaneous melanoma. Mol. Oncol. 2011, 5, 150–155.
- Gradilone, A.; Cigna, E.; Agliano, A.M.; Frati, L. Tyrosinase Expression as a Molecular Marker for Investigating the Presence of Circulating Tumor Cells in Melanoma Patients. Curr. Cancer Drug Tar. 2010, 10, 529–538.
- Jawaid, S.; Khan, T.H.; Osborn, H.M.; Williams, N.A. Tyrosinase activated melanoma prodrugs. Anticancer. Agents Med. Chem. 2009, 9, 717–727.
- Carballo-Carbajal, I.; Laguna, A.; Romero-Gimenez, J.; Cuadros, T.; Bove, J.; Martinez-Vicente, M.; Parent, A.; Gonzalez-Sepulveda, M.; Penuelas, N.; Torra, A.; et al. Brain tyrosinase overexpression implicates age-dependent neuromelanin production in Parkinson’s disease pathogenesis. Nat. Commun. 2019, 10, 973.
- da Silva, G.F.; Ming, L.J. Alzheimer’s disease related copper(II)- beta-amyloid peptide exhibits phenol monooxygenase and catechol oxidase activities. Angew. Chem. Int. Ed. Engl. 2005, 44, 5501–5504.
- Hasegawa, T. Tyrosinase-expressing neuronal cell line as in vitro model of Parkinson’s disease. Int. J. Mol. Sci. 2010, 11, 1082–1089.
- Wang, F.Y.; Wang, P.; Zhao, D.F.; Gonzalez, F.J.; Fan, Y.F.; Xia, Y.L.; Ge, G.B.; Yang, L. Analytical methodologies for sensing catechol-O-methyltransferase activity and their applications. J. Pharm. Anal. 2021, 11, 15–27.
- Cordes, P.; Sun, W.; Wolber, R.; Kolbe, L.; Klebe, G.; Rohm, K.H. Expression in non-melanogenic systems and purification of soluble variants of human tyrosinase. Biol. Chem. 2013, 394, 685–693.
- Dolinska, M.B.; Wingfield, P.T.; Sergeev, Y.V. Purification of Recombinant Human Tyrosinase from Insect Larvae Infected with the Baculovirus Vector. Curr. Protoc. Protein. Sci. 2017, 89, 6.15.1–6.15.12.
- Matoba, Y.; Kumagai, T.; Yamamoto, A.; Yoshitsu, H.; Sugiyama, M. Crystallographic evidence that the dinuclear copper center of tyrosinase is flexible during catalysis. J. Biol. Chem. 2006, 281, 8981–8990.
- Kim, Y.J.; Uyama, H. Tyrosinase inhibitors from natural and synthetic sources: Structure, inhibition mechanism and perspective for the future. Cell Mol. Life Sci. 2005, 62, 1707–1723.
- Zolghadri, S.; Bahrami, A.; Hassan Khan, M.T.; Munoz-Munoz, J.; Garcia-Molina, F.; Garcia-Canovas, F.; Saboury, A.A. A comprehensive review on tyrosinase inhibitors. J. Enzyme Inhib. Med. Chem. 2019, 34, 279–309.
- Ismaya, W.T.; Rozeboom, H.J.; Weijn, A.; Mes, J.J.; Fusetti, F.; Wichers, H.J.; Dijkstra, B.W. Crystal structure of Agaricus bisporus mushroom tyrosinase: Identity of the tetramer subunits and interaction with tropolone. Biochemistry 2011, 50, 5477–5486.
- Ionita, E.; Aprodu, I.; Stanciuc, N.; Rapeanu, G.; Bahrim, G. Advances in structure-function relationships of tyrosinase from Agaricus bisporus-investigation on heat-induced conformational changes. Food Chem. 2014, 156, 129–136.
- Sendovski, M.; Kanteev, M.; Ben-Yosef, V.S.; Adir, N.; Fishman, A. First structures of an active bacterial tyrosinase reveal copper plasticity. J. Mol. Biol. 2011, 405, 227–237.
- Bijelic, A.; Pretzler, M.; Molitor, C.; Zekiri, F.; Rompel, A. The Structure of a Plant Tyrosinase from Walnut Leaves Reveals the Importance of “Substrate-Guiding Residues” for Enzymatic Specificity. Angew. Chem. Int. Ed. Engl. 2015, 54, 14677–14680.
- Fenoll, L.G.; Peñalver, M.J.; Rodríguez-López, J.N.; Varón, R.; García-Cánovas, F.; Tudela, J. Tyrosinase kinetics: Discrimination between two models to explain the oxidation mechanism of monophenol and diphenol substrates. Int. J. Biochem. Cell Biol. 2004, 36, 235–246.
- Fenoll, L.G.; Rodriguez-Lopez, J.N.; Garcia-Sevilla, F.; Garcia-Ruiz, P.A.; Varon, R.; Garcia-Canovas, F.; Tudela, J. Analysis and interpretation of the action mechanism of mushroom tyrosinase on monophenols and diphenols generating highly unstable o-quinones. Biochim. Biophys. Acta 2001, 1548, 1–22.
- Sanchez-Ferrer, A.; Rodriguez-Lopez, J.N.; Garcia-Canovas, F.; Garcia-Carmona, F. Tyrosinase: A comprehensive review of its mechanism. Biochim. Biophys. Acta 1995, 1247, 1–11.
- Qu, Y.; Zhan, Q.; Du, S.; Ding, Y.; Fang, B.; Du, W.; Wu, Q.; Yu, H.; Li, L.; Huang, W. Catalysis-based specific detection and inhibition of tyrosinase and their application. J. Pharm. Anal. 2020, 10, 414–425.
- Rescigno, A.; Sollai, F.; Pisu, B.; Rinaldi, A.; Sanjust, E. Tyrosinase inhibition: General and applied aspects. J. Enzyme Inhib. Med. Chem. 2002, 17, 207–218.
- Sanjust, E.; Cecchini, G.; Sollai, F.; Curreli, N.; Rescigno, A. 3-Hydroxykynurenine as a substrate/activator for mushroom tyrosinase. Arch. Biochem. Biophys 2003, 412, 272–278.
- McLarin, M.A.; Leung, I.K.H. Substrate specificity of polyphenol oxidase. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 274–308.
- Gasowska, B.; Kafarski, P.; Wojtasek, H. Interaction of mushroom tyrosinase with aromatic amines, o-diamines and o-aminophenols. Biochim. Biophys. Acta 2004, 1673, 170–177.
- Rescigno, A.; Bruyneel, F.; Padiglia, A.; Sollai, F.; Salis, A.; Marchand-Brynaert, J.; Sanjust, E. Structure-activity relationships of various amino-hydroxy-benzenesulfonic acids and sulfonamides as tyrosinase substrates. Biochim. Biophys. Acta 2011, 1810, 799–807.
- Espin, J.C.; Garcia-Ruiz, P.A.; Tudela, J.; Garcia-Canovas, F. Study of stereospecificity in mushroom tyrosinase. Biochem. J. 1998, 331, 547–551.
- Espin, J.C.; Varon, R.; Fenoll, L.G.; Gilabert, M.A.; Garcia-Ruiz, P.A.; Tudela, J.; Garcia-Canovas, F. Kinetic characterization of the substrate specificity and mechanism of mushroom tyrosinase. Eur. J. Biochem. 2000, 267, 1270–1279.
- Toussaint, O.; Lerch, K.J.B. Catalytic oxidation of 2-aminophenols and ortho hydroxylation of aromatic amines by tyrosinase. Biochemistry 1987, 26, 8567–8571.
More
Information
Subjects:
Chemistry, Medicinal
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.1K
Revisions:
2 times
(View History)
Update Date:
08 Sep 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




