Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Fabrizio Bianchi | + 1560 word(s) | 1560 | 2021-08-12 09:46:58 | | | |
| 2 | Lily Guo | Meta information modification | 1560 | 2021-08-31 04:18:20 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Bianchi, F.; Colangelo, T. Lung Cancer Biomarkers. Encyclopedia. Available online: https://encyclopedia.pub/entry/13710 (accessed on 07 February 2026).
Bianchi F, Colangelo T. Lung Cancer Biomarkers. Encyclopedia. Available at: https://encyclopedia.pub/entry/13710. Accessed February 07, 2026.
Bianchi, Fabrizio, Tommaso Colangelo. "Lung Cancer Biomarkers" Encyclopedia, https://encyclopedia.pub/entry/13710 (accessed February 07, 2026).
Bianchi, F., & Colangelo, T. (2021, August 30). Lung Cancer Biomarkers. In Encyclopedia. https://encyclopedia.pub/entry/13710
Bianchi, Fabrizio and Tommaso Colangelo. "Lung Cancer Biomarkers." Encyclopedia. Web. 30 August, 2021.
Copy Citation
Lung cancer is the leading cause of cancer death worldwide. Detecting lung malignancies promptly is essential for any anticancer treatment to reduce mortality and morbidity, especially in high-risk individuals.
lung cancer
early diagnosis
1. Introduction
Lung cancer is an aggressive disease accounting for ~380,000 deaths/year only in Europe (WHO; http://gco.iarc.fr; accessed on 21 April 2021) and ~2 million deaths/year worldwide. With the COVID-19 pandemic, these rates are unfortunately expected to rise, mainly due to delays in screening, hospitalizations and therapies, which will cause a stage-shift for newly diagnosed lung tumors [1][2][3].
Detecting lung malignancies promptly is essential for any anticancer treatment to reduce mortality and morbidity, especially in high-risk individuals [4]. The US National Lung Screening Trial (NLST) and other non-randomized trials [5] demonstrated that Low-Dose Computed Tomography (LDCT) screening can reduce mortality (~20%). Recently, the European NELSON trial has observed a lung cancer mortality reduction of ~25% at 10 years and up to ~30% at 10 years [6]. The drawback of LDCT screening is the presence of uncertainties about high costs, risk of radiation exposure, and false positives observed in the screening population [7], which may obstacle a fully safe large scale implementation of the LDCT screening for lung cancer in Europe [8]. The false-positive rate is particularly problematic, as suspicious nodules may require invasive investigations, causing unnecessary morbidity and reduced acceptance of screening among at-risk individuals. Therefore, the integration of LDCT screening with innovative cancer biomarkers analyzable through minimally invasive approaches aimed to increase screening accuracy is highly demanded. Several pre-clinical studies have suggested that circulating molecules such as microRNA, DNA, proteins, autoantibodies in the blood, as well as circulating tumor cells (CTCs), could be potentially useful to diagnose lung cancer and increase screening accuracy [9][10][11][12]. In addition, some studies in actual lung cancer screening cohorts confirmed the diagnostic validity of measuring blood biomarkers for lung cancer early detection [13][14][15]. Yet, pitfalls and caveats emerged during validation of some proposed biomarkers for lung cancer early detection once applied to independent cohorts/multicenter studies and/or actual lung cancer screening cohorts, which highlight the need to establish a roadmap to develop effective biomarkers.
We reviewed the literature for the most promising biomarkers and relevant technical issues, of which here we present a summary with the aim to propose guidelines for an accurate study design and execution, and data interpretation for biomarker development. We hope that these guidelines will aid further research and facilitate the translation of circulating biomarkers into clinical setting.
2. Lung Cancer Biomarkers
In the last 10 years, there has been a sharp rise in published studies on lung cancer diagnostic biomarkers, with over 544 papers published only in the last 5 years (Figure 1A). However, a sizable fraction of these works relies on a relatively small cohort of samples analyzed, without validation of biomarkers in independent cohorts and, more importantly, in lung cancer screening trials. Ideally, robust biomarker(s) should facilitate the selection of at-risk individuals independently of risk factors such as age and smoking habits, and/or provide pathological information about indeterminate pulmonary nodules (IPNs) to aid clinical decision making, and/or provide predictive/prognostic information. Here, we focused on the most promising minimally invasive, reproducible and extensively validated biomarkers assessed in prospective studies, including lung cancer screening trials.
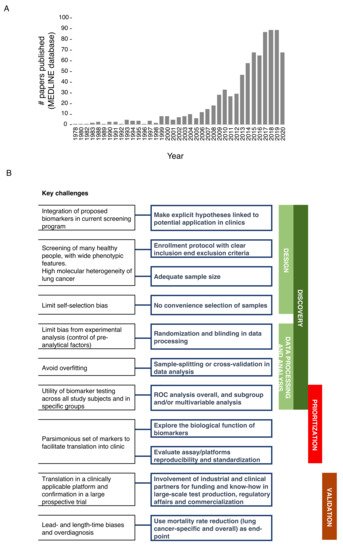
Figure 1. (A) Papers on lung cancer diagnostic biomarkers. PubMed free search engine which primarily accesses the MEDLINE database was interrogated (April 2021) by using ‘advanced search’ tool and with the following MESH terms: Lung neoplasms; Biomarkers; Diagnosis. (B) Schematic representation of best practice in biomarker development for early detection of lung cancer.
3. A Roadmap to the Successful Development of Blood-Based Biomarkers for Lung Cancer Early Detection
The bottleneck for the successful translation of biomarkers to the clinical use generally lies in the suboptimal standardization in each step of the biomarker pipeline, including discovery, prioritization, and clinical validation. We prepared a summary of the main issues and the best practices in biomarker development (Figure 1B). The first fundamental step in biomarker discovery is establishing a high-quality design which includes making explicit hypotheses on the potential application/integration into current recommended screening programs as well as adopting enrollment protocols with clear inclusion and exclusion criteria for patients and controls. Moreover, heterogeneity (epidemiological, biological and molecular) needs to be considered as the driver for adequate sample size to fulfill the best design. Indeed, published studies often lack acceptable sample size with respect to the numerous phenotypic features that should be considered to widely represent the screening population [16], and the number of variables that should be analyzed to deconvolute the high level of genetic heterogeneity of lung cancer. To limit self-selection bias, instead of convenience selection of subjects (based on easy availability of the sample) [17], control populations should be identified based on matching criteria with the patients’ cohort, and extensively represent the actual incidence and prevalence of lung cancer in the screening population.
In the absence of standards for handling specimens (collection, storage and processing) and controls for pre-analytical factors, randomization and blinding should be applied to reduce bias from the experimental analysis. Indeed, quality and reproducibility of biomarkers can be influenced by uncontrolled pre-analytical conditions (i.e., fasting, lipemia, partial hemolysis [18]) and by sample collection bias, especially when the biomarker is labile or sensitive to temperature fluctuation or handling conditions (i.e., type of collection tubes, centrifugation steps, long-term or short-term storage, freeze/thaw cycles; [19][20]). We therefore suggest performing initial pilot experiments to measure the stability of circulating biomarkers, i.e.: (i) by testing different samples collection strategies, using different collection tubes for serum or plasma collection [21][22][23][24]; (ii) quantifying how much hemolysis (partial or hidden) can influence biomarker concentration [25][26], (iii) checking if analyte concentration is influenced by fasting status [18], and (iv) testing if different storage conditions (short-term vs. long-term; +4 or −20/−80 °C or liquid nitrogen) can alter biomarker quantity and quality [18]. After such analyses, a standard operating procedure (SOP) for sample collection and handling should be defined and rigorously applied to the specific biomarkers screening study.
Nowadays, high-throughput data allow the identification of many biomarkers acting jointly on the risk of lung cancer; these markers can be easily combined in a single multivariable statistical model; moreover, to avoid the resulting possible overfitting (i.e., capturing noise instead of the true underlying data structure), machine learning approaches with sample-splitting or cross-validation should be considered [27]. The performance of a new biomarker for the early detection of cancer is easily measured by true-positive and false-positive rates, and summarized through receiver operating characteristic curves (ROC). However, the “average” performance is often presented in the literature, with ROC calculated across all study subjects, while subgroup and/or multivariable analysis should better reveal the utility of biomarker testing in specific groups (i.e., tumor stages, nodule density, histotypes).
Exploration of biomarkers’ performance in subgroups could also help with ranking the selected candidates for clinical relevance. Moreover, when a new biomarker study is published, only limited discussion on the biological function of the candidates is reported, and assay/platform reproducibility and standardization are frequently lacking (see below). In our experience, an in-depth analysis of technical and biological variables which might have an impact on the detection and quantification of selected biomarkers should also be performed. For example, uncontrolled environmental conditions during sample processing could influence the quantification of biomarkers of interest. Marzi et al. [18] showed, by using an automated purification system based on spin columns for nucleic acid purification, that efficiency in miRNA extraction was inversely proportional to temperature increase during daily runs. Similar findings were also described by other research groups [28].
In the case of analysis of multiple biomarkers (e.g., DNA, RNA and protein), the collected samples (whole blood, plasma, serum) can be split in several aliquots which can be differently prioritized for processing based on stability of the biomarkers of interest; in case of RNA, which is more liable, the relevant sample aliquot can be processed immediately while other aliquots (for other biomarker types) can be processed subsequently. Likewise, the use of different extraction kits with or without additional centrifugation steps could affect quantities and species of the biomarkers of interest. Cheng et al. [29] showed that plasma samples can be contaminated by residual platelets, which impact most miRNA measurements (~70%), therefore authors suggested to add pre- or post-storage centrifugation steps in order to remove residual platelet contamination. Furthermore, miRNA quantities may vary depending on the kit used for extraction [30][31].
To keep track of the impact of these pre-analytical and analytical variables, we strongly recommend using endogenous and exogenous controls. In circulating miRNA, biomarker analysis measuring both endogenous controls (e.g., RNU6, RNU44, miR-16 [32]) and exogenous controls, e.g., synthetic miRNAs from other organisms (ath-miR-159a and/or cel-miR-39), allows monitoring sample degradation, extraction efficiency and performance of miRNA detection by using different screening platforms (e.g., qRT-PCR, ddPCR, microarray, NGS).
Lastly, the analytical translation in a clinically applicable platform and validation in a large prospective trial are both needed to complete validation of candidate biomarkers. Industrial and clinical partners could facilitate these phases, providing funding supports and know-how in large-scale test production, regulatory affairs and commercialization [16]. A major issue in the validation of biomarkers for lung cancer early detection is to prove its benefit in the context of screening programs, where lead- and length-time biases and overdiagnosis are peculiar. Therefore, the choice of the end-point is essential and, although biases could occur in interpreting causes of death, lung-cancer mortality reduction should represent the primary endpoint [27], then followed by the evaluation of overall mortality.
References
- Degeling, K.; Baxter, N.N.; Emery, J.; Jenkins, M.A.; Franchini, F.; Gibbs, P.; Mann, G.B.; McArthur, G.; Solomon, B.J.; IJzerman, M.J. An Inverse Stage-Shift Model to Estimate the Excess Mortality and Health Economic Impact of Delayed Access to Cancer Services Due to the COVID-19 Pandemic. Asia Pac. J. Clin. Oncol. 2021.
- Dinmohamed, A.G.; Visser, O.; Verhoeven, R.H.A.; Louwman, M.W.J.; van Nederveen, F.H.; Willems, S.M.; Merkx, M.A.W.; Lemmens, V.E.P.P.; Nagtegaal, I.D.; Siesling, S. Fewer Cancer Diagnoses during the COVID-19 Epidemic in The Netherlands. Lancet Oncol. 2020, 21, 750–751.
- Sud, A.; Jones, M.E.; Broggio, J.; Loveday, C.; Torr, B.; Garrett, A.; Nicol, D.L.; Jhanji, S.; Boyce, S.A.; Gronthoud, F.; et al. Collateral Damage: The impact on outcomes from cancer surgery of the Covid-19 pandemic. Ann. Oncol. 2020, 31, 1065–1074.
- McMahon, P.M.; Kong, C.Y.; Johnson, B.E.; Weinstein, M.C.; Weeks, J.C.; Kuntz, K.M.; Shepard, J.-A.O.; Swensen, S.J.; Gazelle, G.S. Estimating long-term effectiveness of lung cancer screening in the mayo ct screening study. Radiology 2008, 248, 278–287.
- Aberle, D.R.; Adams, A.M.; Berg, C.D.; Black, W.C.; Clapp, J.D.; Fagerstrom, R.M.; Gareen, I.F.; Gatsonis, C.; Marcus, P.M.; Sicks, J.D. Reduced lung-cancer mortality with low-dose computed tomographic screening. New Engl. J. Med. 2011, 365, 395–409.
- De Koning, H.J.; Van Der Aalst, C.M.; De Jong, P.A.; Scholten, E.T.; Nackaerts, K.; Heuvelmans, M.A.; Lammers, J.-W.J.; Weenink, C.; Yousaf-Khan, U.; Horeweg, N.; et al. Reduced lung-cancer mortality with volume ct screening in a randomized trial. New Engl. J. Med. 2020, 382, 503–513.
- Kinsinger, L.S.; Anderson, C.; Kim, J.; Larson, M.; Chan, S.H.; King, H.A.; Rice, K.L.; Slatore, C.G.; Tanner, N.T.; Pittman, K.; et al. Implementation of Lung Cancer Screening in the Veterans Health Administration. JAMA Intern. Med. 2017, 177, 399–406.
- Puggina, A.; Broumas, A.; Ricciardi, W.; Boccia, S. Cost-effectiveness of screening for lung cancer with low-dose computed tomography: A systematic literature review. Eur. J. Public Health 2016, 26, 168–175.
- Bianchi, F.; Nicassio, F.; Marzi, M.; Belloni, E.; Dall’olio, V.; Bernard, L.; Pelosi, G.; Maisonneuve, P.; Veronesi, G.; Di Fiore, P.P. A serum circulating mirna diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol. Med. 2011, 3, 495–503.
- Hofman, V.; Bonnetaud, C.; Ilie, M.I.; Vielh, P.; Vignaud, J.M.; Fléjou, J.F.; Lantuejoul, S.; Piaton, E.; Mourad, N.; Butori, C.; et al. Preoperative circulating tumor cell detection using the isolation by size of epithelial tumor cell method for patients with lung cancer is a new prognostic biomarker. Clin. Cancer Res. 2011, 17, 827–835.
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor dna with broad patient coverage. Nat. Med. 2014, 20, 548–554.
- Tockman, M.S.; Gupta, P.K.; Myers, J.D.; Frost, J.K.; Baylin, S.B.; Gold, E.B.; Chase, A.M.; Wilkinson, P.H.; Mulshine, J.L. Sensitive and specific monoclonal antibody recognition of human lung cancer antigen on preserved sputum cells: A new approach to early lung cancer detection. J. Clin. Oncol. 1988, 6, 1685–1693.
- Montani, F.; Marzi, M.J.; Dezi, F.; Dama, E.; Carletti, R.M.; Bonizzi, G.; Bertolotti, R.; Bellomi, M.; Rampinelli, C.; Maisonneuve, P.; et al. MiR-Test: A blood test for lung cancer early detection. J. Natl. Cancer Inst. 2015, 107.
- Sozzi, G.; Boeri, M.; Rossi, M.; Verri, C.; Suatoni, P.; Bravi, F.; Roz, L.; Conte, D.; Grassi, M.; Sverzellati, N.; et al. Clinical utility of a plasma-based mirna signature classifier within computed tomography lung cancer screening: A correlative mild trial study. J. Clin. Oncol. 2014, 32, 768–773.
- Ajona, D.; Pajares, M.J.; Corrales, L.; Perez-Gracia, J.L.; Agorreta, J.; Lozano, M.D.; Torre, W.; Massion, P.P.; de-Torres, J.P.; Jantus-Lewintre, E.; et al. Investigation of complement activation product c4d as a diagnostic and prognostic biomarker for lung cancer. JNCI J. Natl. Cancer Inst. 2013, 105, 1385–1393.
- Poste, G. Bring on the biomarkers. Nature 2011, 469, 156–157.
- Goossens, N.; Nakagawa, S.; Sun, X.; Hoshida, Y. Cancer biomarker discovery and validation. Transl. Cancer Res. 2015, 4, 256–269.
- Marzi, M.J.; Montani, F.; Carletti, R.M.; Dezi, F.; Dama, E.; Bonizzi, G.; Sandri, M.T.; Rampinelli, C.; Bellomi, M.; Maisonneuve, P.; et al. Optimization and standardization of circulating microRNA detection for clinical application: The miR-test case. Clin. Chem. 2016, 62, 743–754.
- Marton, M.J.; Weiner, R. Practical guidance for implementing predictive biomarkers into early phase clinical studies. BioMed Res. Int. 2013, 2013, 891391.
- Yin, P.; Lehmann, R.; Xu, G. Effects of pre-analytical processes on blood samples used in metabolomics studies. Anal. Bioanal. Chem. 2015, 407, 4879–4892.
- Zhang, S.; Zhao, Z.; Duan, W.; Li, Z.; Nan, Z.; Du, H.; Wang, M.; Yang, J.; Huang, C. The influence of blood collection tubes in biomarkers’ screening by mass spectrometry. Proteom. Clin. Appl. 2020, 14, e1900113.
- Yang, S.; McGookey, M.; Wang, Y.; Cataland, S.R.; Wu, H.M. Effect of blood sampling, processing, and storage on the measurement of complement activation biomarkers. Am. J. Clin. Pathol. 2015, 143, 558–565.
- Risberg, B.; Tsui, D.W.Y.; Biggs, H.; De Almagro, A.R.-V.M.; Dawson, S.-J.; Hodgkin, C.; Jones, L.; Parkinson, C.; Piskorz, A.; Marass, F.; et al. Effects of collection and processing procedures on plasma circulating cell-free DNA from cancer patients. J. Mol. Diagn. 2018, 20, 883–892.
- Glinge, C.; Clauss, S.; Boddum, K.; Jabbari, R.; Jabbari, J.; Risgaard, B.; Tomsits, P.; Hildebrand, B.; Kääb, S.; Wakili, R.; et al. Stability of circulating blood-based microRNAs—Pre-analytic methodological considerations. PLoS ONE 2017, 12, e0167969.
- Poel, D.; Buffart, T.E.; Oosterling-Jansen, J.; Verheul, H.M.; Voortman, J. Evaluation of several methodological challenges in circulating miRNA qPCR studies in patients with head and neck cancer. Exp. Mol. Med. 2018, 50, e454.
- Kirschner, M.B.; Edelman, J.J.; Kao, S.C.; Vallely, M.P.; Van Zandwijk, N.; Reid, G. The impact of hemolysis on cell-free microRNA Biomarkers. Front. Genet. 2013, 4, 94.
- Maruvada, P. Joint national cancer institute-food and drug administration workshop on research strategies, study designs, and statistical approaches to biomarker validation for cancer diagnosis and detection. Cancer Epidemiol. Biomark. Prev. 2006, 15, 1078–1082.
- Sourvinou, I.S.; Markou, A.; Lianidou, E.S. Quantification of circulating miRNAs in plasma. J. Mol. Diagn. 2013, 15, 827–834.
- Cheng, H.H.; Yi, H.S.; Kim, Y.; Kroh, E.M.; Chien, J.W.; Eaton, K.D.; Goodman, M.T.; Tait, J.F.; Tewari, M.; Pritchard, C.C. Plasma processing conditions substantially influence circulating microRNA biomarker levels. PLoS ONE 2013, 8, e64795.
- El-Khoury, V.; Pierson, S.; Kaoma, T.; Bernardin, F.; Berchem, G. Assessing cellular and circulating miRNA recovery: The impact of the RNA isolation method and the quantity of input material. Sci. Rep. 2016, 6, 19529.
- Kloten, V.; Neumann, M.H.D.; Di Pasquale, F.; Sprenger-Haussels, M.; Shaffer, J.M.; Schlumpberger, M.; Herdean, A.; Betsou, F.; Ammerlaan, W.; Hällström, T.; et al. Multicenter evaluation of circulating plasma microRNA extraction technologies for the development of clinically feasible reverse transcription quantitative PCR and next-generation sequencing analytical work flows. Clin. Chem. 2019, 65, 1132–1140.
- Schwarzenbach, H.; Silva, A.M.; Calin, G.; Pantel, K. Data normalization strategies for microRNA quantification. Clin. Chem. 2015, 61, 1333–1342.
More
Information
Subjects:
Oncology; Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
685
Revisions:
2 times
(View History)
Update Date:
02 Sep 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




