Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Susana Santos Lopes | + 2659 word(s) | 2659 | 2021-08-11 08:39:49 | | | |
| 2 | Rita Xu | Meta information modification | 2659 | 2021-08-17 03:18:04 | | | | |
| 3 | Conner Chen | Meta information modification | 2659 | 2021-09-22 03:46:51 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Santos Lopes, S. Zebrafish Motile Cilia. Encyclopedia. Available online: https://encyclopedia.pub/entry/13227 (accessed on 07 February 2026).
Santos Lopes S. Zebrafish Motile Cilia. Encyclopedia. Available at: https://encyclopedia.pub/entry/13227. Accessed February 07, 2026.
Santos Lopes, Susana. "Zebrafish Motile Cilia" Encyclopedia, https://encyclopedia.pub/entry/13227 (accessed February 07, 2026).
Santos Lopes, S. (2021, August 16). Zebrafish Motile Cilia. In Encyclopedia. https://encyclopedia.pub/entry/13227
Santos Lopes, Susana. "Zebrafish Motile Cilia." Encyclopedia. Web. 16 August, 2021.
Copy Citation
Zebrafish is a vertebrate teleost widely used in many areas of research. As embryos, they develop quickly and provide unique opportunities for research studies owing to their transparency for at least 48 h post fertilization. Zebrafish have many ciliated organs that include primary cilia as well as motile cilia. Using zebrafish as an animal model helps to better understand human diseases such as Primary Ciliary Dyskinesia (PCD), an autosomal recessive disorder that affects cilia motility, currently associated with more than 50 genes.
transmission electron microscopy
motile cilia
left–right organizer
1. Introduction
Motile cilia are centriole-derived organelles, surrounded by a membrane and containing microtubules formed by protofilaments that can be longer or shorter, depending on the number of tubulin molecules they contain, according to the function they perform [1][2][3]. This structure is called the ciliary axoneme or ciliary shaft [4], a transversal term throughout species, due to the well-conserved ciliary structure [3]. The axonemal ultrastructure in eukaryotic cells segregates into two significant patterns: 9 + 2, in which nine microtubule doublets organise around two microtubules in the centre known as the central pair (CP) complex; the 9 + 0 organisation, in which the CP is absent [5][6]. Moreover, there are two major types of motile ciliated cells: cells that produce from dozens to several hundred 9 + 2 motile cilia and cells that only generate one cilium.
Motile monocilia-lacking CPs were described in the node of mice [7][8][9]. These are very different from non-motile primary cilia (also with a 9 + 0 configuration) without dynein motor arms that can be found in almost every cell type having solo sensory functions and lacking the ability to generate movement [10][11]. On the other hand, Yu et al. in 2011 identified cilia with a 9 + 2 organisation but immotile, called kinocilia, in the hair cells of the inner ear that express Foxj1b [12] and in the ciliated receptor cells (primary sensory cells) of the zebrafish OP [13]. We can, thus, speculate that all combinations are present in nature, likely by adaptive evolution.
Regarding motile cilia dynamics, multiciliated cells that typically have cilia with a 9 + 2 configuration beat metachronically with a planar stroke to clear fluid or promote locomotion [14][15]. On the other hand, motile cilia lacking CP (9 + 0) have a specific movement pattern, described as planar rotation, circling or twisting. These 9 + 0 monocilia are present in the LRO in vertebrates such as mice or zebrafish [16][17][18], but reports on LRO cilia with 9 + 2 cilia and 9 + 4 also exist [7][18][19]. In cases of disease, multiciliated cells with cilia wholly or partially lacking the CP are frequently accounted as one of the phenotypes of Primary Ciliary Dyskinesia, an autosomal recessive disease of the motile cilia [20][21]. The rationale being that in the respiratory cilia, a rotational movement is not efficient in mucociliary clearance and is, therefore, causative of PCD symptoms.
Zebrafish became popular as an animal model in the 1980s, triggered by George Streisinger’s studies showing zebrafish as a genetically tractable organism, allowing a phenotypic characterisation of a large number of mutations that cause defects in a variety of organ systems [22]. As zebrafish are vertebrates, the translational interpretation of ciliary defects is very powerful, when compared, for instance, to Chlamydomonas reinhardtii, where the defects from cilia impairment are usually spotted as a lack of locomotion [23], although not exclusively. Zebrafish helped shed light on the role of specific genes in human diseases, as their genome has been widely studied, providing insight into their human orthologues [24]. Amongst the many advantages of zebrafish, we will stress the fact that zebrafish embryos contain cilia in nearly every cell type, and their organogenesis defects can be easily characterised using brightfield and fluorescent microscopy as the zebrafish embryo and larvae are mostly transparent until all major organ systems are formed. Many of the organs and tissues of zebrafish are similar to those of humans and 70% of genes are shared [24]. Motile cilia have been well described in the Kupffer’s vesicle, the zebrafish LRO, between 3 and 14 somite stages (ss) of development and in the OP at 48–72 hpf, amongst other locations [25].
Zebrafish, as almost all teleosts, have three types of olfactory receptor neurons (ORNs): ciliated ORNs, microvillous ORNs and crypt cells with both cilia and microvilli [26][27]. The bottom of the OP is coated with ORNs, each one with protruding non-motile primary cilia that contain olfactory receptors [28]. On the other hand, the rim is surrounded by a layer of cuboidal multiciliated cells protruding bundles of motile cilia [28]. These microvillous ORNs, and ciliated ORNs have similar morphological and molecular contents to the microvillous and ciliated ORNs of higher vertebrates [29]. Moreover, genetic engineering methodologies, including CRISPR/Cas9, have been successfully used in editing the zebrafish genome and have greatly facilitated the generation of zebrafish mutant models mimicking human ciliopathies [30][31][32].
PCD is a genetic disorder distinguished by recurrent infection in the lower and upper respiratory tract [33], reduced fertility and laterality problems (50% of PCD patients show situs inversus) [34]. Ultrastructural defects in motile cilia or a reduced cilia number are known to cause PCD [34][35]. Currently there are more than 50 genes identified that can cause PCD [36]. Almost all PCD genes show homologous genes in zebrafish, as shown in Table 1. Despite previous research on zebrafish cilia [13][18][26][37], no study fully characterised the cilia ultrastructure of the OP and LRO. With this work, we aim to determine the similarities and the significant differences between zebrafish and human motile cilia, comparing OP multiciliated cells and embryonic LRO monociliated cells. Using transmission electron microscopy (TEM), we confirmed the heterogenic configuration of the zebrafish OP cells. We also showed by electron tomography (ET) the variable presence of a CP in the LRO monocilia, a feature hypothesised in a previous publication by Tavares et al. [18].
Table 1. Human-Zebrafish homologue PCD genes as described in Ensemble.org [38].
| PCD Gene | Zebrafish Transcript Name | Zebrafish Transcript ID |
|---|---|---|
| DNAH5 | dnah5-201 | ENSDART00000123150.4 |
| dnah5-202 | ENSDART00000191818.1 | |
| CCDC114 (ODAD1) | ccdc114-201 | ENSDART00000023745.8 |
| ARMC4 (ODAD2) | cr847789.1-201 | ENSDART00000186851.1 |
| armc4-201 | ENSDART00000077453.5 | |
| armc4-204 | ENSDART00000170018.2 | |
| armc4-203 | ENSDART00000153115.2 | |
| armc4-202 | ENSDART00000152887.2 | |
| TTC25 (ODAD4) | ttc25-201 | ENSDART00000080946.5 |
| DNAH9 | dnah9-201 | ENSDART00000160926.2 |
| DNAH11 | dnah11-201 | ENSDART00000148294.4 |
| dnah11-202 | ENSDART00000020821.10 | |
| dnah11-203 | ENSDART00000138744.4 | |
| DNAI1 | dnai1.2-201 | ENSDART00000080431.5 |
| dnai1.2-202 | ENSDART00000142468.3 | |
| dnai1.1-205 | ENSDART00000170205.2 | |
| dnai1.1-201 | ENSDART00000160163.2 | |
| dnai1.1-204 | ENSDART00000169676.2 | |
| dnai1.1-202 | ENSDART00000163063.2 | |
| dnai1.1-203 | ENSDART00000165798.2 | |
| DNAI2 | dnai2a-201 | ENSDART00000162579.2 |
| dnai2a-202 | ENSDART00000164199.2 | |
| dnai2b-203 | ENSDART00000188726.1 | |
| dnai2b-201 | ENSDART00000003339.9 | |
| dnai2b-202 | ENSDART00000188648.1 | |
| DNAL1 | dnal1-203 | ENSDART00000188500.1 |
| dnal1-202 | ENSDART00000156182.2 | |
| dnal1-201 | ENSDART00000043651.7 | |
| TXNDC3 (NME8) | nme8-201 | ENSDART00000163684.2 |
| CCDC103 | ccdc103-201 | ENSDART00000075493.4 |
| ccdc103-202 | ENSDART00000132293.2 | |
| CFAP298 (C21orf59) | cfap298-201 | ENSDART00000051197.6 |
| cfap298-202 | ENSDART00000130093.3 | |
| cfap298-203 | ENSDART00000181950.1 | |
| CFAP300 (c11orf70) | cfap300-201 | ENSDART00000151109.2 |
| cfap300-202 | ENSDART00000192737.1 | |
| DNAAF1 (LRRC50) | dnaaf1-201 | ENSDART00000145762.4 |
| dnaaf1-203 | ENSDART00000173909.2 | |
| dnaaf1-202 | ENSDART00000173853.2 | |
| DNAAF2 (KTU) | dnaaf2-201 | ENSDART00000167840.2 |
| DNAAF3 | dnaaf3l-201 | ENSDART00000079233.5 |
| DNAAF4 (DYX1C1) | dnaaf4-201 | ENSDART00000165855.2 |
| DNAAF5 (HEATR2) | lo018183.1-201 | ENSDART00000194031.1 |
| DNAAF6 (PIH1D3) | pih1d3-201 | ENSDART00000056375.5 |
| pih1d3-203 | ENSDART00000145388.3 | |
| pih1d3-202 | ENSDART00000136858.2 | |
| pih1d3-204 | ENSDART00000183524.1 | |
| pih1d3-205 | ENSDART00000191761.1 | |
| LRRC6 | lrrc6-203 | ENSDART00000188883.1 |
| lrrc6-202 | ENSDART00000132346.3 | |
| lrrc6-201 | ENSDART00000075347.5 | |
| RPGR | rpgrb-201 | ENSDART00000088624.5 |
| rpgrb-202 | ENSDART00000124471.3 | |
| rpgrip-201 | ENSDART00000138541.3 | |
| rpgrip-203 | ENSDART00000190953.1 | |
| rpgrip-202 | ENSDART00000179003.2 | |
| rpgrip1l-202 | ENSDART00000185324.1 | |
| rpgrip1l-201 | ENSDART00000126326.5 | |
| SPAG1 | spag1b-201 | ENSDART00000101207.5 |
| spag1a-202 | ENSDART00000185960.1 | |
| spag1a-201 | ENSDART00000130537.3 | |
| ZMYND10 | zmynd10-201 | ENSDART00000017413.10 |
| zmynd10-202 | ENSDART00000189261.1 | |
| zmynd10-203 | ENSDART00000183251.1 | |
| CCDC39 | ccdc39-202 | ENSDART00000190769.1 |
| ccdc39-201 | ENSDART00000169709.2 | |
| CCDC40 | ccdc40-202 | ENSDART00000169752.2 |
| Ccdc40-201 | ENSDART00000164275.2 | |
| Ccdc40-203 | ENSDART00000182267.1 | |
| TTC12 | ttc12-201 | ENSDART00000156234.2 |
| ttc12-202 | ENSDART00000157380.2 | |
| CCDC65 (DRC2) | ccdc65-201 | ENSDART00000043946.8 |
| ccdc65-202 | ENSDART00000177219.2 | |
| CCDC164 (DRC1) | drc1-201 | ENSDART00000061829.5 |
| GAS8 | gas8-202 | ENSDART00000170982.2 |
| gas8-201 | ENSDART00000165126.2 | |
| CFAP221 | not found in ZF | |
| DNAJB13 | dnajb13-204 | ENSDART00000148093.3 |
| dnajb13-201 | ENSDART00000063365.6 | |
| dnajb13-203 | ENSDART00000139097.2 | |
| dnajb13-202 | ENSDART00000133505.2 | |
| HYDIN | hydin-201 | ENSDART00000143265.4 |
| hydin-202 | ENSDART00000145701.2 | |
| hydin-203 | ENSDART00000169861.2 | |
| bx571975.1-201 | ENSDART00000185269.1 | |
| NME5 | nme5-201 | ENSDART00000060998.6 |
| RSPH1 | rsph1-201 | ENSDART00000160273.3 |
| ct573248.3-201 | ENSDART00000181186.1 | |
| RSPH3 | rsph3-202 | ENSDART00000128823.5 |
| rsph3-201 | ENSDART00000103394.3 | |
| RSPH4a | rsph4a-201 | ENSDART00000097340.5 |
| RSPH9 | rsph9-201 | ENSDART00000010903.8 |
| STK36 | stk36-201 | ENSDART00000086765.5 |
| stk36-202 | ENSDART00000139065.2 | |
| SPEF2 | spef2-201 | ENSDART00000159718.2 |
| spef2-202 | ENSDART00000168984.2 | |
| CFAP57 | cfap57-201 | ENSDART00000080900.6 |
| cfap57-202 | ENSDART00000149309.3 | |
| LRRC56 | lrrc56-202 | ENSDART00000161369.2 |
| lrrc56-201 | ENSDART00000150364.2 | |
| GAS2L2 | gas2l2-201 | ENSDART00000112744.4 |
| NEK10 | nek10-201 | ENSDART00000155162.2 |
| OFD1 | ofd1-201 | ENSDART00000000552.12 |
| CCNO | fq311924.1-201 | ENSDART00000158096.2 |
| FOXJ1 | foxj1b-201 | ENSDART00000126676.2 |
| foxj1b-203 | ENSDART00000181942.1 | |
| foxj1b-202 | ENSDART00000153327.2 | |
| foxj1a-201 | ENSDART00000157772.2 | |
| foxj1a-202 | ENSDART00000168280.2 | |
| MCIDAS | cu633857.1-201 | ENSDART00000192716.1 |
2. Zebrafish Motile Cilia
Zebrafish have motile cilia in many of its organs, that are present since the early stages of development, as depicted in the diagram from Figure 1.
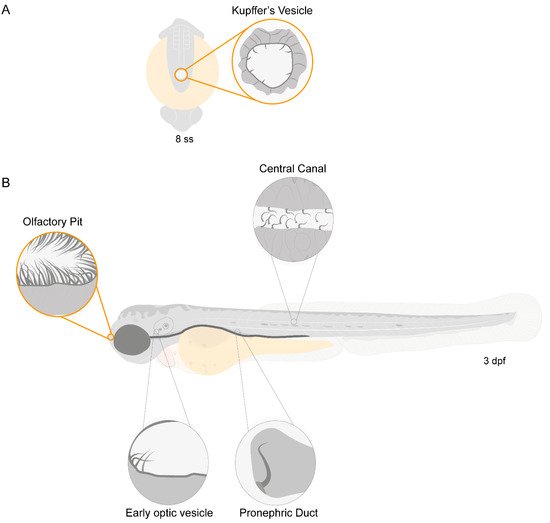
Figure 1. Motile ciliated structures in zebrafish. Schematic representation of (A), a zebrafish embryo at 8 somite stages (ss) highlighting Kupffer’s vesicle (KV); (B) a 3-day post-fertilization (dpf) zebrafish larva indicating structures with motile cilia. Olfactory pit (OP) and KV cilia analysed in the present study are circled in orange.
Olstad et al. have previously shown the flow generated by the OP motile cilia [39]. Whereas Sampaio et al. (2014) and Tavares et al. (2017) have extensively described the rotational and wavy fashion beat of monocilia of the LRO [16][18]. These motile monocilia are essential to generate flow for the determination of left–right asymmetry [16][18][40]. We confirmed the localization and distribution of cilia in the wild-type (WT) LRO and OP of zebrafish by immunofluorescence (IF). To better understand how to orient the embryo for the subsequent TEM embedding and sectioning, we generated 3D blend projections and surfaces as shown in Figure 2 for the OP. We consider that this study greatly helped the TEM work. Confocal microscopy is useful but limited to 180 nm lateral and 500 nm axial resolution and is not appropriate for ultrastructural studies. Therefore, we next used TEM to access the ultrastructure of the respective organ cilia.
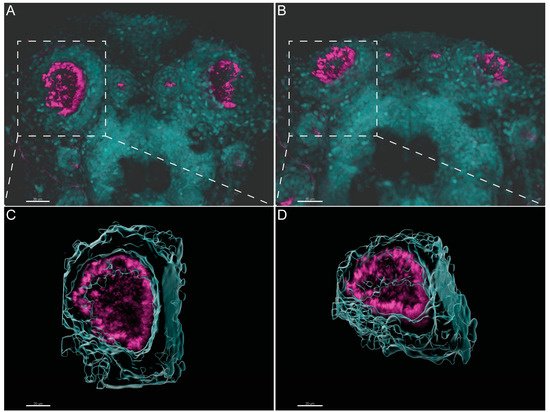
Figure 2. Three-dimensional imaging analysis helps to orient and localize the OP of zebrafish for TEM studies. (A,B). Immunofluorescent labelling with anti-acetylated α-tubulin shows the distribution of multiciliated cells in the OP of 4 dpf larvae. Software Imaris (Bitplane) v.9.5.0 allowed 3D blend reconstructions of 2 different OPs from 2 different larvae. (C,D) 3D surface reconstructions from the respective OPs revealing the concave morphology of the organ when rotated. Anti-Acetylated α-tubulin immunofluorescence in magenta and DAPI in cyan. Scale bar 20 μm.
Next, we evaluated the cilia beat frequency for both organs, so that, in the future, researchers can compare it with zebrafish disease models for PCD. CBF in the LRO ranges from 15 to 50 Hz was evaluated by CiliarMove [41]. This software creates a heatmap for CBF that allowed us to unequivocally detect in a very visual way different cilia in the same focal plane beating differently as depicted by the blue, green and yellow ciliary colour codes (Figure 3A’). This raised an intriguing question as to what is the function of this spatial heterogeneity in the LRO CBF? Compared to the OP, where all cilia beat around 20 Hz, in the green colour code (Figure 3B’), it is tempting to speculate that monocilia in the LRO do not coordinate their CBF for a reason that may relate to the establishment of asymmetry. In addition to the intra-LRO cilia CBF variability, we also detected an inter-embryonic variability greater in the LRO than in the OP (Figure 3C).
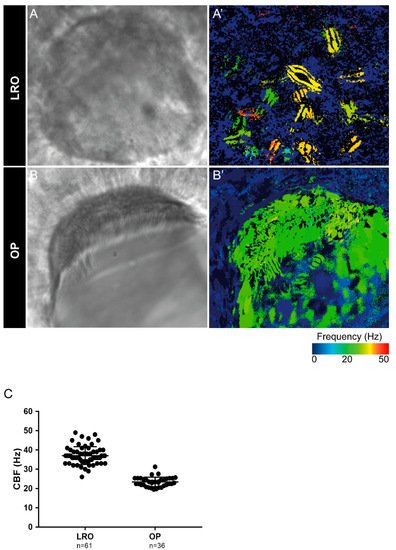
Figure 3. Cilia beat frequency (CBF) evaluation for zebrafish cilia. (A,A’) Full LRO heatmap showing several monocilia at one plane beating at different frequencies (20–50 Hz). (B,B’) OP heatmap showing multiciliated cells with cilia beating at more homogeneous frequencies (around 20 Hz). CBF was measured using the software CiliarMove [41] (C) Quantification of CBF from n = 61 LROs and n = 36 OPs from embryos at 10 ss and 4 dpf larvae, respectively.
3. Different Cilia in the Zebrafish Olfactory Pit
To characterise the ultrastructural pattern of cilia in the OP, a detailed investigation was conducted by means of examining cilia from five dpf zebrafish by TEM. The quantitative analysis of three different WT animals (N = 113 cilia; values show mean ± standard deviation) showed that 60% (± 1) of cilia had a 9 + 2 arrangement with dynein arms present, and 23% (± 2) of cilia presented absent or incomplete dynein arms (most notably in the outer dynein arm (ODA)) (Figure 4 and Table 2). We further showed the heterogenicity of cilia within specific regions of the OP. When determining the localisation of the different ciliary ultrastructural types, cilia observed in the peripheral OP had dynein arms reflecting their motile function, whereas cilia in the central OP had absence of ODA and inner dynein arms (IDA). A TEM analysis allowed the quantification of the heterogenicity of cilia in a specific region of the OP. Cilia observed in the OP had a 9 + 2 motile morphology in a more peripheral area and a 9 + 2 with an ODA and IDA absence towards the centre of the OP.
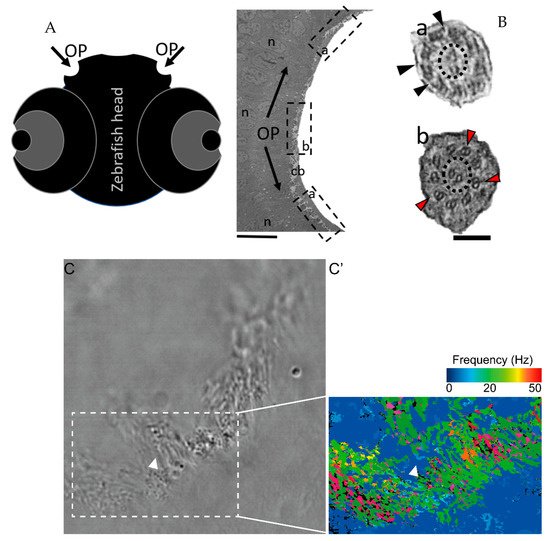
Figure 4. The zebrafish OP has two different types of cilia. (A) Schematic representation of 5 dpf zebrafish head, structures of interest are marked with arrows—olfactory pits (OP). (B) TEM low magnification image showing a cross-section across an OP of a WT zebrafish, a bowl-shaped structure containing multiciliated cells and cilia in several orientations. In the periphery of this pit (dotted boxes a and inset), cilia containing classical motile structure 9 + 2 with dynein arms (black arrowheads). In the most internal region of the OP (dotted box b and inset) we detected cilia with 9 + 2 ultrastructural arrangement without dynein arms (red arrowheads); this pattern was visible in WT embryos (n = 3). (C) Snapshot from a movie of beating OP cilia and respective heatmap by software CiliarMove (C’), highlighting a region of immotile cilia (arrowhead) that was coincident with the region detected by TEM in B. n—nucleus; cb—cilia border; OP—olfactory pit. Thin bar 6 µm, thick bar 100 nm.
Table 2. Full TEM assessment and quantification of defects in cilia from the OP in wildtype zebrafish and human healthy controls. N = 3 zebrafish OPs and n = 113 cilia were examined, values shown are mean ± standard deviation.
| Disarranged Cilia (%) | Dynein Arms Assessment (%) | ||||
|---|---|---|---|---|---|
| Counted > 50 Cilia | Both Arms Present | ODA Missing | IDA Missing | Both Arms Missing | |
| WT zebrafish (n = 3) | 14 (±8) | 62 (±1) | 23 (±2) | 14 (±2) | 14 (±2) |
| Human control (n = 3) | 3 (±1) | 93 (±12) | 1 (±1) | 3 (±6) | 2 (±4) |
The percentages of missing ODA and IDA assessed by quantitative methods [42] (as shown in Table 2) were compared using Student’s t-test in both zebrafish and human samples for a significance analysis. The comparisons between WT zebrafish OP cilia and healthy control patients concerning the presence of ODAs and IDAs were significantly different (ODA *** p ≤ 0.001; IDA * p ≤ 0.05, Table 2 and Figure 5). In healthy humans, ODAs were rarely missing, contrary to the observed findings in the central region of the zebrafish OP. Therefore, for research purposes of modelling PCD using zebrafish OP cilia, one should consider cilia from the OP periphery.
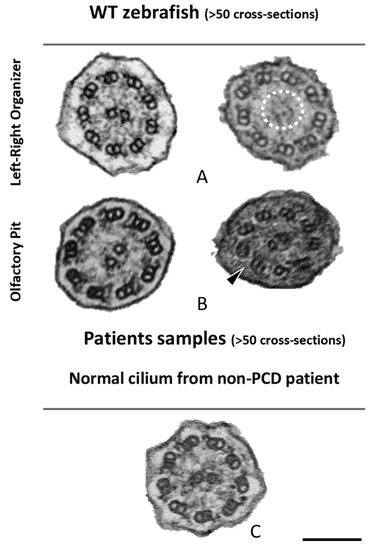
Figure 5. Ciliary ultrastructural comparison between the LRO, OP and human respiratory cilia. Variations of the axonemal arrangement of cilia in (A) the LRO of 10 somite stages WT zebrafish embryo and (B) the OP of 5 dpf WT zebrafish larvae. (C) Example of a human respiratory cilium cross-section from the airway. Arrowheads indicate missing ODA and the dotted circle shows missing CP. Scale bar 100 nm.
Next, cross-sections from the LRO cilia were also analysed by TEM (Figure 5). A limited number of cilia were observed due to the monociliated nature of the LRO cells. The ultrastructure of these cilia, as shown previously [18], had dynein arms in WT embryos (n > 100 cilia) and some cilia show an absent or partial configuration of the CP as shown in Figure 5.
To compare the structure of the microtubule doublets of zebrafish LRO cilia against human respiratory cilia, we performed ET to generate 3D reconstructions. After assessing the cilia from the LRO by ET, some concern regarding the size of the ODA was raised, as some variation in the size of the outer dynein arms was visible, suggesting it might be smaller. To clarify, we used the software Chimera [43] (USFC, California) to measure the volume of the ODA which was normalised to the total volume of the microtubule doublet (MTD). We analysed the volume of the MTD from the LRO of four different WT zebrafish and compared them to the MTD of three human control respiratory cilia (Figure 6). No significant differences were found between the two samples, indicating that LRO cilia and human airway cilia have similar ODA volumes (student’s test, p > 0.05).
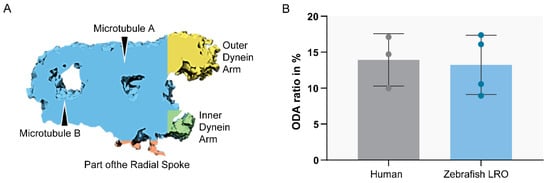
Figure 6. Ratio between ODA and MTD volume. Sub-tomographic averaging of the MTD of WT zebrafish cilia, showing ODA and IDA, tubules (A,B) and a portion of the radial spoke. The two sample groups were not significantly different (p > 0.05, Student’s t-test). N = 3 human patients and n = 4 zebrafish LROs.
References
- Nicastro, D.; Schwartz, C.; Pierson, J.; Gaudette, R.; Porter, M.E.; McIntosh, J.R. The molecular architecture of axonemes revealed by cryoelectron tomography. Science 2006, 313, 944–948.
- Ishikawa, T. Axoneme structure from motile cilia. Cold Spring Harb. Perspect. Biol. 2017, 9, a028076.
- Vincensini, L.; Blisnick, T.; Bastin, P. 1001 Model Organisms to Study Cilia and Flagella. Biol. Cell 2011, 103, 109–130.
- Legendre, M.; Zaragosi, L.E.; Mitchison, H.M. Motile cilia and airway disease. In Seminars in Cell & Developmental Biology; Elsevier: London, UK, 2020.
- Porter, K.R. The submicroscopic morphology of protoplasm. 1956. Anat. Rec. A. Discov. Mol. Cell. Evol. Biol. 2005, 287, 1186–1204.
- Satir, P.; Christensen, S.T. Overview of Structure and Function of Mammalian Cilia. Annu. Rev. Physiol. 2007, 69, 377–400.
- Caspary, T.; Larkins, C.E.; Anderson, K.V. The Graded Response to Sonic Hedgehog Depends on Cilia Architecture. Dev. Cell 2007, 12, 767–778.
- Nonaka, S.; Shiratori, H.; Saijoh, Y.; Hamada, H. Determination of left-right patterning of the mouse embryo by artificial nodal flow. Nature 2002, 418, 96–99.
- Nonaka, S.; Tanaka, Y.; Okada, Y.; Takeda, S.; Harada, A.; Kanai, Y.; Kido, M.; Hirokawa, N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 1998, 95, 829–837.
- Gluenz, E.; Höög, J.L.; Smith, A.E.; Dawe, H.R.; Shaw, M.K.; Gull, K. Beyond 9 + 0: Noncanonical axoneme structures characterize sensory cilia from protists to humans. FASEB J. 2010, 24, 3117–3121.
- Sánchez, I.; Dynlacht, B.D. Cilium assembly and disassembly. Nat. Cell Biol. 2016, 18, 711–717.
- Yu, X.; Lau, D.; Ng, C.P.; Roy, S. Cilia-driven fluid flow as an epigenetic cue for otolith biomineralization on sensory hair cells of the inner ear. Development 2011, 138, 487–494.
- Hansen, A.; Zeiske, E. The Peripheral Olfactory Organ of the Zebrafish, Danio Rerio: An Ultrastructural Study; Oxford University Press: Oxford, UK, 1998; Volume 23.
- Del Bigio, M.R. Ependymal cells: Biology and pathology. Acta Neuropathol. 2010, 119, 55–73.
- Satir, P.; Sleigh, M.A. The physiology of cilia and mucociliary interactions. Annu. Rev. Physiol. 1990, 52, 137–155.
- Sampaio, P.; Ferreira, R.R.; Guerrero, A.; Pintado, P.; Tavares, B.; Amaro, J.; Smith, A.A.; Montenegro-Johnson, T.; Smith, D.J.; Lopes, S.S. Article Left-Right Organizer Flow Dynamics: How Much Cilia Activity Reliably Yields Laterality? Dev. Cell 2014, 29, 716–728.
- Roxo-Rosa, M.; Santos Lopes, S. The Zebrafish Kupffer’s Vesicle: A Special Organ in a Model Organism to Study Human Diseases. In Zebrafish in Biomedical Research; American College of Laboratory Animal Medicina series; USA IntechOpen: New York, NY, USA, 2020.
- Tavares, B.; Jacinto, R.; Sampaio, P.; Pestana, S.; Pinto, A.; Vaz, A.; Roxo-Rosa, M.; Gardner, R.; Lopes, T.; Schilling, B.; et al. Notch/Her12 signalling modulates, motile/immotile cilia ratio downstream of Foxj1a in zebrafish left-right organizer. Elife 2017, 6, e25165.
- Feistel, K.; Blum, M. Three types of cilia including a novel 9 + 4 axoneme on the notochordal plate of the rabbit embryo. Dev. Dyn. 2006, 235, 3348–3358.
- Yokeid, H.; Ueno, H.; Narita, A.; Sakai, T.; Horiuchiid, K.; Shingyojiid, C.; Hamada, H.; Shinoharaid, K. Rsph4a is essential for the triplet radial spoke head assembly of the mouse motile cilia. PLoS Genet. 2020, 16, e1008664.
- Castleman, V.H.; Romio, L.; Chodhari, R.; Hirst, R.A.; de Castro, S.C.P.; Parker, K.A.; Ybot-Gonzalez, P.; Emes, R.D.; Wilson, S.W.; Wallis, C.; et al. Mutations in radial spoke head protein genes RSPH9 and RSPH4A cause primary ciliary dyskinesia with central-microtubular-pair abnormalities. Am. J. Hum. Genet. 2008, 84, 197–209.
- Driever, W.; Solnica-Krezel, L.; Schier, A.F.; Neuhauss, S.C.F.; Malicki, J.; Stemple, D.L.; Stainier, D.Y.R.; Zwartkruis, F.; Abdelilah, S.; Rangini, Z.; et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development 1996, 123, 37–46.
- Liu, G.; Wang, L.; Pan, J.; Yao, X. Chlamydomonas WDR92 in association with R2TP-like complex and multiple DNAAFs to regulate ciliary dynein preassembly. J. Mol. Cell Biol. 2019, 11, 770–780.
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503.
- Drummond, I.; Austin-Tse, C. Zebrafish cilia. In Methods in Enzymology; Elsevier Group: London, UK, 2013; Volume 525, pp. 219–244. ISBN 9780123979445.
- Hansen, A.; Zielinski, B.S. Diversity in the olfactory epithelium of bony fishes: Development, lamellar arrangement, sensory neuron cell types and transduction components. J. Neurocytol. 2006, 208, 183–208.
- Olivares, J.; Schmachtenberg, O. An update on anatomy and function of the teleost olfactory system. PeerJ 2019, 7, 1–20.
- Neuhauss, S.C.F. Olfaction: How Fish Catch a Whiff. Curr. Biol. 2017, 27, R57–R58.
- Hansen, A.; Rolen, S.H.; Anderson, K.; Morita, Y.; Caprio, J.; Finger, T.E. Correlation between Olfactory Receptor Cell Type and Function in the Channel Catfish. J. Neurosci. 2003, 23, 9328–9339.
- Song, Z.; Zhang, X.; Jia, S.; Yelick, P.C.; Zhao, C. Zebrafish as a Model for Human Ciliopathies. J. Genet. Genom. 2016, 43, 107–120.
- Liu, D.; Wang, Z.; Xiao, A.; Zhang, Y.; Li, W.; Zu, Y.; Yao, S.; Lin, S.; Zhang, B. Efficient Gene Targeting in Zebrafish Mediated by a Zebrafish-Codon-Optimized Cas9 and Evaluation of Off-Targeting Effect. J. Genet. Genom. 2014, 41, 43–46.
- Chang, N.; Sun, C.; Gao, L.; Zhu, D.; Xu, X.; Zhu, X.; Xiong, J.W.; Xi, J.J. Genome editing with RNA-guided Cas9 nuclease in Zebrafish embryos. Cell Res. 2013, 23, 465–472.
- Kouis, P.; Yiallouros, P.K.; Middleton, N.; Evans, J.S.; Kyriacou, K.; Papatheodorou, S.I. Prevalence of primary ciliary dyskinesia in consecutive referrals of suspect cases and the transmission electron microscopy detection rate: A systematic review and meta-analysis. Pediatr. Res. 2016, 81, 398–405.
- Lucas, J.S.; Burgess, A.; Mitchison, H.M.; Moya, E.; Williamson, M.; Hogg, C. Diagnosis and management of primary ciliary dyskinesia. Arch. Dis. Child. 2014, 99, 850–856.
- Mitchison, H.M.; Valente, E.M. Motile and non-motile cilia in human pathology: From function to phenotypes. J. Pathol. 2017, 241, 294–309.
- Wallmeier, J.; Nielsen, K.G.; Kuehni, C.E.; Lucas, J.S.; Leigh, M.W.; Zariwala, M.A.; Omran, H. Motile ciliopathies. Nat. Rev. Dis. Prim. 2020, 6, 1–29.
- Reiten, I.; Uslu, F.E.; Fore, S.; Pelgrims, R.; Ringers, C.; Diaz Verdugo, C.; Hoffman, M.; Lal, P.; Kawakami, K.; Pekkan, K.; et al. Motile-Cilia-Mediated Flow Improves Sensitivity and Temporal Resolution of Olfactory Computations. Curr. Biol. 2017, 27, 166–174.
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688.
- Olstad, E.W.; Ringers, C.; Hansen, J.N.; Wens, A.; Brandt, C.; Wachten, D.; Yaksi, E.; Jurisch-Yaksi, N. Ciliary Beating Compartmentalizes Cerebrospinal Fluid Flow in the Brain and Regulates Ventricular Development. Curr. Biol. 2019, 29, 229–241.e6.
- Dasgupta, A.; Amack, J.D. Cilia in vertebrate left—Right patterning. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150410.
- Sampaio, P.; da Silva, M.F.; Vale, I.; Roxo-Rosa, M.; Pinto, A.; Constant, C.; Pereira, L.; Quintão, C.M.; Lopes, S.S. CiliarMove: New software for evaluating ciliary beat frequency helps find novel mutations by a Portuguese multidisciplinary team on primary ciliary dyskinesia. ERJ Open Res. 2021, 7, 00792–02020.
- Pinto, A.L.; Rai, R.K.; Hogg, C.; Burgoyne, T. Ciliary feature counter: A program for the quantitative assessment of cilia to diagnose primary ciliary dyskinesia. Diagnostics 2020, 10, 524.
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612.
More
Information
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.4K
Revisions:
3 times
(View History)
Update Date:
22 Sep 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




