Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Lijuan Tan | + 2997 word(s) | 2997 | 2021-08-04 08:52:37 | | | |
| 2 | Bruce Ren | -21 word(s) | 2976 | 2021-08-12 11:00:55 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Tan, L. CNN in Leaf Disease Classification. Encyclopedia. Available online: https://encyclopedia.pub/entry/13083 (accessed on 09 March 2026).
Tan L. CNN in Leaf Disease Classification. Encyclopedia. Available at: https://encyclopedia.pub/entry/13083. Accessed March 09, 2026.
Tan, Lijuan. "CNN in Leaf Disease Classification" Encyclopedia, https://encyclopedia.pub/entry/13083 (accessed March 09, 2026).
Tan, L. (2021, August 12). CNN in Leaf Disease Classification. In Encyclopedia. https://encyclopedia.pub/entry/13083
Tan, Lijuan. "CNN in Leaf Disease Classification." Encyclopedia. Web. 12 August, 2021.
Copy Citation
Crop production can be greatly reduced due to various diseases, which seriously endangers food security. Thus, detecting plant diseases accurately is necessary and urgent. Traditional classification methods, such as naked-eye observation and laboratory tests, have many limitations, such as being time consuming and subjective. Currently, deep learning (DL) methods, especially those based on convolutional neural network (CNN), have gained widespread application in plant disease classification. They have solved or partially solved the problems of traditional classification methods and represent state-of-the-art technology in this field.
plant disease classification
deep learning
machine learning
convolutional neural network
1. Introduction
The Food and Agriculture Organization of the United Nations (http://www.fao.org/publications/sofi/2020/en/, accessed on 5 December 2020) reported that the number of hungry people in the world has been increasing slowly since 2014. Current estimates show that nearly 690 million people are hungry, and they account for 8.9% of the world’s total population; this figure represents an increase of 10 million in 1 year and nearly 60 million in 5 years. Meanwhile, more than 90% of people in the world rely on agriculture. Farmers produce 80% of the world’s food [1]; however, more than 50% of crop production is lost due to plant diseases and pests [2]. Thus, recognizing and detecting plant disease accurately is necessary and urgent.
The diverse plant diseases have an enormous effect on growing food crops. An iconic example is the Irish potato famine of 1845–1849, which resulted in 1.2 million deaths [3]. The diseases of several common plants are shown in Table 1. Plant diseases can be systematically divided into fungal, oomycete, hyphomycete, bacterial, and viral types. We have shown some pictures of plant disease in Figure 1. The pictures in Figure 1 were taken in the greenhouse of Chengdu Academy of Agriculture and Forestry Sciences. Researchers and farmers have never stopped exploring how to develop an intelligent and effective method for plant disease classification. Laboratory test approaches to plant samples, such as polymerase chain reaction, enzyme-linked immunosorbent assay, and loop-mediated isothermal amplification, are highly specific and sensitive in identifying diseases.

Figure 1. Leaf spot in eight common plants. We took these pictures in the greenhouse of Chengdu Academy of Agriculture and Forestry Sciences.
Table 1. Common diseases of several common plants.
| Plant | Major Types of Disease | Reference | ||
|---|---|---|---|---|
| Fungal | Bacterial | Viral | ||
| Cucumber | Downy mildew, powdery mildew, gray mold, black spot, anthracnose | Angular spot, brown spot, target spot | Mosaic virus, yellow spot virus | Kianat et al. (2021) [4], Zhang et al. (2019) [5], Agarwal et al. (2021) [6] |
| Rice | Rice stripe blight, false smut, rice blast | Bacterial leaf blight, bacterial leaf streak | Rice leaf smut, rice black-streaked dwarf virus | Chen et al. (2021) [7], Shrivastava et al. (2019) [8] |
| Maize | Leaf spot disease, rust disease, gray leaf spot | Bacterial stalk rot, bacterial leaf streak | Rough dwarf disease, crimson leaf disease | Sun et al. (2021) [9], Yu et al. (2014) [10] |
| Tomato | Early blight, late blight, leaf mold | Bacterial wilt, soft rot, canker | Tomato yellow leaf curl virus | Ferentinos (2018) [11], Abbas et al. (2021) [12] |
However, conventional field scouting for diseases in crops still relies primarily on visual inspection of the leaf color patterns and crown structures. People observe the symptoms of diseases on plant leaves with the naked eye and diagnose plant diseases based on experience, which is time and labor consuming and requires specialized skills [13]. At the same time, the disease characteristics among different crops are also different due to the variety of plants; this condition brings a high degree of complexity in the classification of plant diseases. Meanwhile, many studies have focused on the classification of plant diseases based on machine learning. Using machine learning methods to detect plant diseases is mainly divided into the following three steps: first, using preprocessing techniques to remove the background or segment the infected part; second, extracting the distinguishing features for further analysis; finally, using supervised classification or unsupervised clustering algorithms to classify the features [14][15][16][17]. Most machine learning studies have focused on the classification of plant diseases by using features, such as the texture [18], type [19], and color [20] of plant leaf images. The main classification methods include support vector machines [19], K-nearest neighbor [20], and random forest [21]. The major disadvantages of these methods are summarized as follows:
Low performance [22]: The performance they obtained was not ideal and could not be used for real-time classification.
Professional database [23]: The datasets they applied contained plant images that were difficult to obtain in actual life. In the case of PlantVillage, the dataset was taken in an ideal laboratory environment, such that a single image contains only one plant leaf and the shot is not influenced by the external environment (e.g., light, rain).
Rarely used [24][25]: They often need to manually design and extract features, which require research staff to possess professional capabilities.
Requiring the use of segmented operation [26]: The plants must be separated from their roots to gain research datasets. Obviously, this operation is not good for real-time applications.
Most of the traditional machine learning algorithms were based on laboratory conditions, and the robustness of the algorithms is insufficient to meet the needs of practical agricultural applications. Nowadays, deep learning (DL) methods, especially those based on convolutional neural networks (CNNs), are gaining widespread application in the agricultural field for detection and classification tasks, such as weed detection [27], crop pest classification, and plant disease identification [28]. DL is a research direction of machine learning. It has solved or partially solved the problems of low performance [22], lack of actual images [23], and segmented operation [26] of traditional machine learning methods. The important advantage of DL models are that they can extract features without applying segmented operation while obtaining satisfactory performance. Features of an object are automatically extracted from the original data. Kunihiko Fukushima introduced the Neocognitron in 1980, which inspired CNNs [29]. The emergence of CNNs has made the technology of plant disease classification increasingly efficient and automatic.
The main works of this study are given as follows: (1) we reviewed the latest CNN networks pertinent to plant leaf disease classification; (2) we summarized DL principles involved in plant disease classification; (3) we summarized the main problems and corresponding solutions of CNN used for plant disease classification, and (4) we discussed the direction of future developments in plant disease classification.
2. Deep Learning
DL is a branch of machine learning [30] and is mainly used for image classification, object detection [31][32][33][34], and natural language processing [35][36][37].
DL is an algorithm based on a neural network for automatic feature selection of data. It does not need a lot of artificial feature engineering. It combines low-level features to form abstract high-level features for discovering distributed features and attributes of sample data. Its accuracy and generalization ability are improved compared to those of traditional methods in image recognition and target detection. Currently, the main types of networks are multilayer perceptron, CNN, and recurrent neural network (RNN). CNN is the most widely used for plant leaf disease classification. As for other DL networks, such as fully convolutional networks (FCNs) and deconvolutional networks, they are usually used for image segmentation [38][39][40][41] or medical diagnosis [42][43] but are not used for plant leaf disease classification. CNN usually consists of convolutional, pooling, and fully connected layers. The convolutional layer uses the local correlation of the information in the image to extract features. The process of convolution operation is shown in Figure 2. A kernel is placed in the top-left corner of the image. The pixel values covered by the kernel are multiplied with the corresponding kernel values, and then the products are summated, and the bias is added at the end. The kernel is moved over by one pixel, and the process is repeated until all possible locations in the image are filtered, which is shown in Figure 2. The pooling layer selects features from the upper layer feature map by sampling and simultaneously makes the model invariant to translation, rotation, and scaling. The commonly used one is maximum or average pooling. The process of the pooling operation is shown in Figure 3. Maximum pooling is to divide the input image into several rectangular regions based on the size of the filter and output the maximum value for each region. As for average pooling, the output is the average of each region. Convolutional and pooling layers often appear alternately in applications. Each neuron in the fully connected layer is connected to the upper neuron, and the multidimensional features are integrated and converted into one-dimensional features in the classifier for classification or detection tasks [44].
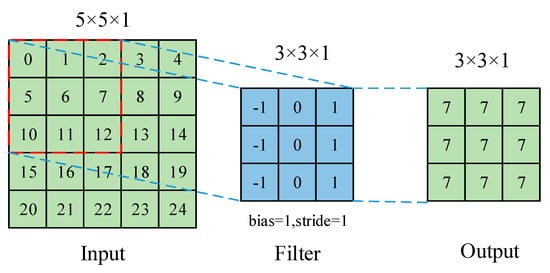
Figure 2. The process of convolution operation.
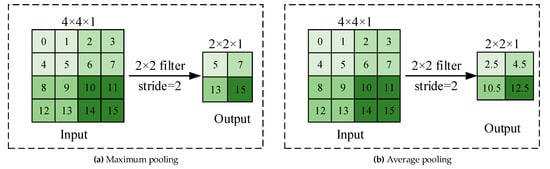
Figure 3. The process of pooling operation.
For classification tasks, various CNN-based classification models have been developed in DL-related research, including AlexNet, VGGNet, GoogLeNet, ResNet, MobileNet, and EfficientNet. AlexNet [45] was proposed in 2012 and was the champion network in the ILSVRC-2012 competition. This network contains five convolutional layers and three fully connected layers. AlexNet has the following four highlights: (a) it is the first model to use a GPU device for network acceleration training; (b) rectified linear units (ReLUs) were used as the activation function; (c) local response normalization was used; (d) in the first two layers of the fully connected layer, the dropout operation was used to reduce overfitting. Then, the deeper networks appeared, such as VGG16, VGG19, GoogLeNet. These networks use smaller stacked kernels but have lower memory during inference [46]. Later, researchers found that when the number of layers of a deep CNN reached a certain depth, blindly increasing the number of layers would not improve the classification performance but would cause the network to converge more slowly [47][48]. Until 2015, Microsoft lab proposed the ResNet network and won the first place in the classification task of the ImageNet competition. The network creatively proposed residual blocks and shortcut connections [49], which solves the problem of gradient elimination or gradient explosion, making it possible to build a deeper network model. ResNet influenced the development direction of DL in academia and industry in 2016. MobileNet was proposed by the Google teams in 2017 and was designed for mobile and embedded vision applications [50]. In 2019, the Google teams proposed another outstanding network: EfficientNet [51]. This network uses a simple yet highly efficient compound coefficient to uniformly scale all dimensions of depth/width/resolution, which will not arbitrarily scale the dimensions of the network as in traditional methods. As for plant disease classification tasks, it is not necessary to use deep networks, because simple models, such as AlexNet and VGG16, can meet the actual accuracy requirements.
The DL model can be realized using programming languages, such as Python, C/C++. The open-source DL framework provides a series of application programming interfaces, supports model design, assists in network deployment, and avoids code duplication [52]. At present, DL frameworks, such as PyTorch (https://pytorch.org/, accessed on 5 March 2021), Tensorflow (https://www.tensorflow.org/, accessed on 7 March 2021), Cafe (https://caffe.berkeleyvision.org/, accessed on 8 March 2021), and Keras (https://keras.io/, accessed on 10 March 2021) are widely used.
The rapid increase of DL is inseparable from the widespread development of GPU. The implementation of deep CNN requires GPUs to provide computing power support, otherwise it will cause the training process to be quite slow or make it impossible to train CNN models at all. At present, the most used is CUDA. When NVIDIA launched CUDA (Computing Unified Device Architecture) and AMD launched Stream, GPU computing started [46], and now, CUDA is widely used in DL.
Image classification is a basic task in computer vision. It is also the basis of object detection, image segmentation, image retrieval, and other technologies. The basic process of DL is shown in Figure 4, taking the task of classification of diseases on the surface of snake gourd leaves as an example. In Figure 4, we use a CNN-based architecture to extract features, which mainly include convolutional, max-pooling, and full connection layers. The convolutional layer is mainly used to extract features of snake gourd plant leaf images. The shallow convolutional layer is used to extract some edge and texture information, the middle layer is used to extract complex texture and part of semantic information, and the deep layer is used to extract high-level semantic features. The convolutional layer is followed by a max-pooling layer, which is used to retain the important information in the image. At the end of the architecture is a classifier, which consists of full connection layers. This classifier is used to classify the high-level semantic features extracted by the feature extractor.
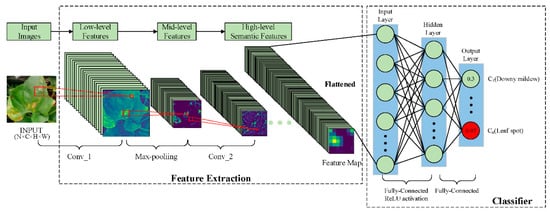
Figure 4. Convolutional neural networks for snake gourd leaf disease classification.
In Figure 4, we input a batch of images into the feature extraction network to extract the features and then flatten the feature map into the classifier for disease classification. This process can be roughly divided into the following three steps.
-
Step 1. Preparing the Data and Preprocessing
-
Step 2. Building, Training, and Evaluating the Model
-
Step 3. Inference and Deployment
2.1. Data Preparation and Preprocessing
Data are important for DL models. The results are bound to be inaccurate no matter how complex and perfect our model is as long as the quality of the input data is poor. The typical percentages of the original dataset intended for training, validation, and test are 70:20:10, 80:10:10, and 60:20:20.
A DL dataset is usually composed of a training set, a validation set, and a test set. The training set is used to make the model learn, and the validation set is usually used to adjust hyperparameters during training. The test set is the sample of data that the model has not seen before, and it is used to evaluate the performance of the DL model. We collected some public plant datasets from the two websites Kaggle (https://www.kaggle.com/datasets, accessed on 12 February 2021) and BIFROST (https://datasets.bifrost.ai/, accessed on 15 February 2021), which can be used for detection or classification tasks, as shown in Table 2. In the literature of DL techniques applied to plant disease classification, the most used public datasets are PlantVillage [53][54][55] and Kaggle [56]; notably, many authors also collect their own datasets [57][58][59][60].
Table 2. Some public plant datasets from Kaggle and BIFROST.
| Name | Number of Images | Classes | Task | Type of View | Source |
|---|---|---|---|---|---|
| New Plant Diseases Dataset | 87,000 | 38 | Image classification | Field data | Kaggle |
| PlantVillage Dataset | 162,916 | 38 | Image classification | Uniform background | Kaggle |
| Flowers Recognition | 4242 | 4 | Image classification | Field data | Kaggle |
| Plant Seedings Dataset | 5539 | 12 | Target detection | Field data | BIFROST |
| Weed Detection in Soybean Crops | 15,336 | 4 | Target detection | Uniform background | Kaggle |
For snake gourd leaf disease classification, we need a large number of leaf images of different disease categories. Meanwhile, the disease image data of each category were roughly balanced. If one disease with a particularly large number of image data is considered, then the neural network will be biased toward this disease. Apart from sufficient data on category balance, it also needs data to preprocess including image resize, random crop, and normalization. The shape of the data varies according to the framework used. Figure 5 shows the tensor shape of the input for the neural network, where H and W represent the height and width of the preprocessed image, C represents the number of image channels (gray or RGB), and N represents the number of images input to the neural network in a training session.
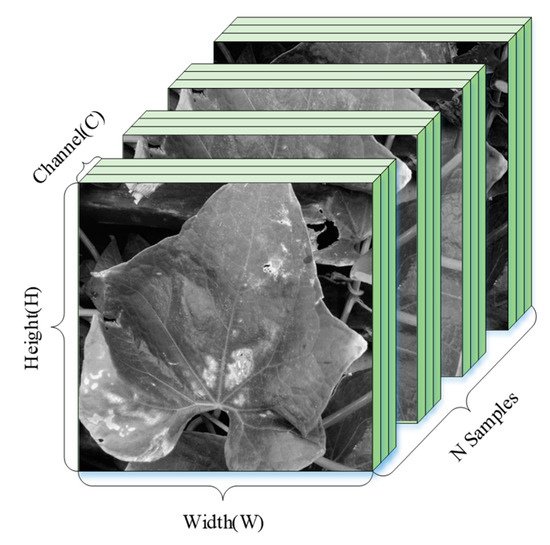
Figure 5. The tensor shape of the input neural network in PyTorch.
2.2. Building Model Architecture, Training, and Evaluating the Model
Before training, a suitable DL model architecture is needed. A good model architecture can result in more accurate classification results and more rapid classification speed. Currently, the main network types of DL are CNN, RNN, and generative adversarial networks (GAN). Among various works, CNN is the most widely used feature extraction network for the task of plant disease detection and classification [55][61][62][63][64][65].
After the model architecture is established, different hyperparameters are set for training and evaluation. We can set some parameter combinations and use the grid search method to iterate through them to find the best one. When training the neural network, training data are placed into the first layer of the network, and each neuron updates the weight of the neuron through back-propagation according to whether the output is equal to the label. This process is repeated until new capability is learned from existing data. However, whether the trained model has learned new capabilities is unknown. The performance of the model was evaluated by criteria, such as accuracy, precision, recall, and F1 score. The concept of a confusion matrix must be introduced first prior to introducing these indexes specifically. The confusion matrix shows the predicted correct or incorrect results in binary classification. It consists of four elements: true positive (TP, correctly predicted positive values), false positive (FP, incorrectly predicted positive values), true negative (TN, correctly predicted negative values), and false negative (FN, incorrectly predicted negative values). Then, the accuracy can be calculated as follows:
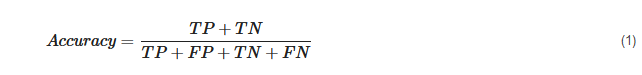
Among all the positives predicted by the model, precision predicts the proportion of correct predictions.

Among all real positives, recall predicts the correct proportion of positives [66].

The F1 value considers precision (P) and recall (R) rates.

In the studies on plant disease classification, accuracy is the most common evaluation index [53][60][64][67][68]. Larger values of accuracy, precision, and recall are better. Within a certain range, when the value of the F1 score is smaller, the better the generalization performance of the trained model is. When the training and evaluation are complete, the trained model has a new capability; then, this capability is applied to new data.
2.3. Inference and Deployment
The inference is the capability of the DL model to quickly apply the learning capability by the trained model to new data and quickly provide the correct answer based on data that it has never seen [69]. After the training process is completed, the networks are deployed into the field for inferring a result for the provided data, which they have never seen before. Only then can the trained deep learning models be applied in real agricultural environments. We can deploy the trained model to the mobile terminal, cloud, or edge devices, such as by using an application on the mobile phone to take photos of plant leaves and judge diseases [70]. In addition, in order to use the trained model better in the field, the generalization ability of the model needs to be improved, and we can continuously update the models with the new labeled datasets to improve the generalization ability [71].
References
- Mukti, I.Z.; Biswas, D. Transfer Learning Based Plant Diseases Detection Using ResNet50. In Proceedings of the 2019 4th International Conference on Electrical Information and Communication Technology (EICT), Khulna, Bangladesh, 20–22 December 2019.
- Arunnehru, J.; Vidhyasagar, B.S.; Anwar Basha, H. Plant Leaf Diseases Recognition Using Convolutional Neural Network and Transfer Learning. In International Conference on Communication, Computing and Electronics Systems; Bindhu, V., Chen, J., Tavares, J.M.R.S., Eds.; Springer: Singapore, 2020; pp. 221–229.
- Hughes, D.P.; Salathe, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv 2015, arXiv:1511.08060.
- Kianat, J.; Khan, M.A.; Sharif, M.; Akram, T.; Rehman, A.; Saba, T. A joint framework of feature reduction and robust feature selection for cucumber leaf diseases recognition. Optik 2021, 240, 166566.
- Zhang, S.; Zhang, S.; Zhang, C.; Wang, X.; Shi, Y. Cucumber leaf disease identification with global pooling dilated convolutional neural network. Comput. Electron. Agric. 2019, 162, 422–430.
- Agarwal, M.; Gupta, S.; Biswas, K.K. A new conv2d model with modified relu activation function for identification of disease type and severity in cucumber plant. Sustain. Comput. Inform. Syst. 2021, 30, 100473.
- Chen, J.; Zhang, D.; Zeb, A.; Nanehkaran, Y.A. Identification of rice plant diseases using lightweight attention networks. Expert Syst. Appl. 2021, 169, 114514.
- Shrivastava, V.K.; Pradhan, M.K.; Minz, S.; Thakur, M.P. Rice plant disease classification using transfer learning of deep convolution neural network. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2019, XLII-3/W6, 631–635.
- Sun, H.; Zhai, L.; Teng, F.; Li, Z.; Zhang, Z. qRgls1. 06, a major QTL conferring resistance to gray leaf spot disease in maize. Crop. J. 2021, 9, 342–350.
- Yu, C.; Ai-hong, Z.; Ai-Jun, R.; Hong-qin, M. Types of Maize virus diseases and progress in virus identification techniques in China. J. Northeast Agric. Univ. 2014, 21, 75–83.
- Ferentinos, K. Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 2018, 145, 311–318.
- Abbas, A.; Jain, S.; Gour, M.; Vankudothu, S. Tomato plant disease detection using transfer learning with C-GAN synthetic images. Comput. Electron. Agric. 2021, 187, 106279.
- Sankaran, S.; Mishra, A.; Ehsani, R.; Davis, C. A review of advanced techniques for detecting plant diseases. Comput. Electron. Agric. 2010, 72, 1–13.
- Barbedo Arnal, J.G. An Automatic Method to Detect and Measure Leaf Disease Symptoms Using Digital Image Processing. Plant Dis. 2014, 98, 1709–1716.
- Feng, Q.; Dongxia, L.; Bingda, S.; Liu, R.; Zhanhong, M.; Haiguang, W. Identification of Alfalfa Leaf Diseases Using Image Recognition Technology. PLoS ONE 2016, 11, e168274.
- Omrani, E.; Khoshnevisan, B.; Shamshirband, S.; Saboohi, H.; Anuar, N.B.; Nasir, M.H.N.M. Potential of radial basis function-based support vector regression for apple disease detection. Measurement 2014, 55, 512–519.
- Barbedo Arnal, J.G. A new automatic method for disease symptom segmentation in digital photographs of plant leaves. Eur. J. Plant Pathol. 2017, 147, 349–364.
- Springer. SVM-Based Detection of Tomato Leaves Diseases; Springer: Berlin/Heidelberg, Germany, 2015.
- Rumpf, T.; Mahlein, A.K.; Steiner, U.; Oerke, E.C.; Dehne, H.W.; Plümer, L. Early detection and classification of plant diseases with Support Vector Machines based on hyperspectral reflectance. Comput. Electron. Agric. 2010, 74, 91–99.
- Hossain, E.; Hossain, M.F.; Rahaman, M.A. A Color and Texture Based Approach for the Detection and Classification of Plant Leaf Disease Using KNN Classifier. In Proceedings of the 2019 International Conference on Electrical, Computer and Communication Engineering (ECCE), Cox’s Bazar, Bangladesh, 7–9 February 2019.
- Mohana, R.M.; Reddy CK, K.; Anisha, P.R.; Murthy, B.R. Random forest algorithms for the classification of tree-based ensemble. Mater. Today Proc. 2021.
- Türkoğlu, M.; Hanbay, D. Plant disease and pest detection using deep learning-based features. Turk. J. Electr. Eng. Comput. Sci. 2019, 27, 1636–1651.
- Arivazhagan, S.; Shebiah, R.N.; Ananthi, S.; Varthini, S.V. Detection of unhealthy region of plant leaves and classification of plant leaf diseases using texture features. Agric. Eng. Int. CIGR J. 2013, 15, 211–217.
- Jiang, F.; Lu, Y.; Chen, Y.; Cai, D.; Li, G. Image recognition of four rice leaf diseases based on deep learning and support vector machine. Comput. Electron. Agric. 2020, 179, 105824.
- Gao, J.; French, A.P.; Pound, M.P.; He, Y.; Pridmore, T.P.; Pieters, J.G. Deep convolutional neural networks for image-based Convolvulus sepium detection in sugar beet fields. Plant Methods 2020, 16, 29.
- Athanikar, G.; Badar, P. Potato leaf diseases detection and classification system. Int. J. Comput. Sci. Mob. Comput. 2016, 5, 76–88.
- Yu, J.; Sharpe, S.M.; Schumann, A.W.; Boyd, N.S. Deep learning for image-based weed detection in turfgrass. Eur. J. Agron. 2019, 104, 78–84.
- Bansal, P.; Kumar, R.; Kumar, S. Disease Detection in Apple Leaves Using Deep Convolutional Neural Network. Agriculture 2021, 11, 617.
- Wang, H.; Raj, B. On the origin of deep learning. arXiv 2017, arXiv:1702.07800.
- Deng, L.; Yu, D. Deep Learning: Methods and Applications. Found. Trends Signal Process. 2014, 7, 197–387.
- Dyrmann, M.; Karstoft, H.; Midtiby, H.S. Plant species classification using deep convolutional neural network. Biosyst. Eng. 2016, 151, 72–80.
- Kussul, N.; Lavreniuk, M.; Skakun, S.; Shelestov, A. Deep Learning Classification of Land Cover and Crop Types Using Remote Sensing Data. IEEE Geosci. Remote. Sens. Lett. 2017, 14, 778–782.
- Wason, R. Deep learning: Evolution and expansion. Cogn. Syst. Res. 2018, 52, 701–708.
- Geetharamani, G.; Pandian, A. Identification of plant leaf diseases using a nine-layer deep convolutional neural network. Comput. Electr. Eng. 2019, 76, 323–338.
- Huang, G.; Liu, Z.; Laurens, V.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. Available online: https://openaccess.thecvf.com/content_cvpr_2017/papers/Huang_Densely_Connected_Convolutional_CVPR_2017_paper.pdf (accessed on 22 July 2021).
- Too, E.C.; Yujian, L.; Njuki, S.; Yingchun, L. A comparative study of fine-tuning deep learning models for plant disease identification. Comput. Electron. Agric. 2018, 161, 272–279.
- He, K.; Zhang, X.; Ren, S.; Jian, S. Identity Mappings in Deep Residual Networks. In European Conference on Computer Vision; Springer: Cham, Switzerland, 2016.
- Guan, S.; Kamona, N.; Loew, M. Segmentation of Thermal Breast Images Using Convolutional and Deconvolutional Neural Networks. In Proceedings of the 2018 IEEE Applied Imagery Pattern Recognition Workshop (AIPR), Washington, DC, USA, 9–11 October 2018.
- Fakhry, A.; Zeng, T.; Ji, S. Residual Deconvolutional Networks for Brain Electron Microscopy Image Segmentation. IEEE Trans. Med. Imaging 2017, 36, 447–456.
- Liu, J.; Wang, Y.; Li, Y.; Fu, J.; Li, J.; Lu, H. Collaborative Deconvolutional Neural Networks for Joint Depth Estimation and Semantic Segmentation. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 5655–5666.
- Wang, J.; Wang, Z.; Tao, D.; See, S.; Wang, G. Learning Common and Specific Features for RGB-D Semantic Segmentation with Deconvolutional Networks. In European Conference on Computer Vision; Springer: Cham, Switzerland, 2016.
- Gehlot, S.; Gupta, A.; Gupta, R. SDCT-AuxNet: DCT Augmented Stain Deconvolutional CNN with Auxiliary Classifier for Cancer Diagnosis. Med. Image Anal. 2020, 61, 101661.
- Duggal, R.; Gupta, A.; Gupta, R.; Mallick, P. SD-Layer: Stain Deconvolutional Layer for CNNs in Medical Microscopic Imaging; Springer: Cham, Switzerland, 2017.
- Gu, J.; Wang, Z.; Kuen, J.; Ma, L.; Wang, G. Recent Advances in Convolutional Neural Networks. Pattern Recognit. 2015, 77, 354–377.
- Krizhevsky, A.; Sutskever, I.; Hinton, G. ImageNet Classification with Deep Convolutional Neural Networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105.
- Gao, Z.; Luo, Z.; Zhang, W.; Lv, Z.; Xu, Y. Deep Learning Application in Plant Stress Imaging: A Review. AgriEngineering 2020, 2, 430–446.
- Bengio, Y.; Simard, P.; Frasconi, P. Learning long-term dependencies with gradient descent is difficult. IEEE Trans. Neural Netw. 1994, 5, 157–166.
- Glorot, X.; Bengio, Y. Understanding the difficulty of training deep feedforward neural networks. J. Mach. Learn. Res. 2010, 9, 249–256.
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2016, arXiv:1512.03385.
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861.
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. arXiv 2019, arXiv:1704.04861.
- Parvat, A.; Chavan, J.; Kadam, S.; Dev, S.; Pathak, V. A survey of deep-learning frameworks: 2017 International Conference on Inventive Systems and Control (ICISC). In Proceedings of the 2017 International Conference on Inventive Systems and Control (ICISC), Coimbatore, India, 19–20 January 2017.
- Mohanty, S.P.; Hughes, D.P.; Marcel, S. Using Deep Learning for Image-Based Plant Disease Detection. Front. Plant Sci. 2016, 7, 1419.
- Brahimi, M.; Mahmoudi, S.; Boukhalfa, K.; Moussaoui, A. Deep interpretable architecture for plant diseases classification. In Proceedings of the 2019 Signal Processing: Algorithms, Architectures, Arrangements, and Applications (SPA), Poznan, Poland, 18–20 September 2019.
- Mishra, S.; Sachan, R.; Rajpal, D. Deep Convolutional Neural Network based Detection System for Real-time Corn Plant Disease Recognition. Procedia Comput. Ence 2020, 167, 2003–2010.
- Darwish, A.; Ezzat, D.; Hassanien, A.E. An optimized model based on convolutional neural networks and orthogonal learning particle swarm optimization algorithm for plant diseases diagnosis. Swarm Evol. Comput. 2019, 52, 100616.
- Amanda, R.; Kelsee, B.; Peter, M.C.; Babuali, A.; James, L.; Hughes, D.P. Deep Learning for Image-Based Cassava Disease Detection. Front. Plant Sci. 2017, 8, 1852.
- Lin, Z.; Mu, S.; Shi, A.; Pang, C.; Student, G.; Sun, X.; Student, G. A Novel Method of Maize Leaf Disease Image Identification Based on a Multichannel Convolutional Neural Network. Trans. ASABE 2018, 61, 1461–1474.
- Yuwana, R.S.; Suryawati, E.; Zilvan, V.; Ramdan, A.; Fauziah, F. Multi-Condition Training on Deep Convolutional Neural Networks for Robust Plant Diseases Detection. In Proceedings of the 2019 International Conference on Computer, Control, Informatics and its Applications (IC3INA), Tangerang, Indonesia, 23–24 October 2019.
- Chen, J.; Chen, J.; Zhang, D.; Sun, Y.; Nanehkaran, Y.A. Using deep transfer learning for image-based plant disease identification. Comput. Electron. Agric. 2020, 173, 105393.
- Fujita, E.; Kawasaki, Y.; Uga, H.; Kagiwada, S.; Iyatomi, H. Basic investigation on a robust and practical plant diagnostic system. In Proceedings of the 2016 15th IEEE International Conference on Machine Learning and Applications (ICMLA), Anaheim, CA, USA, 18–20 December 2016.
- Ghazi, M.M.; Yanikoglu, B.; Aptoula, E. Plant identification using deep neural networks via optimization of transfer learning parameters. Neurocomputing 2017, 235, 228–235.
- Hidayatuloh, A.; Nursalman, M.; Nugraha, E. Identification of Tomato Plant Diseases by Leaf Image Using Squeezenet Model. In Proceedings of the 2018 International Conference on Information Technology Systems and Innovation (ICITSI), Bandung, Indonesia, 22–26 October 2018.
- Juncheng, M.; Keming, D.; Feixiang, Z.; Lingxian, Z.; Zhihong, G.; Zhongfu, S. A recognition method for cucumber diseases using leaf symptom images based on deep convolutional neural network. Comput. Electron. Agric. 2018, 154, 18–24.
- Bollis, E.; Pedrini, H.; Avila, S. Weakly Supervised Learning Guided by Activation Mapping Applied to a Novel Citrus Pest Benchmark. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Seattle, WA, USA, 14–19 June 2020.
- Ge, M.; Su, F.; Zhao, Z.; Su, D. Deep Learning Analysis on Microscopic Imaging in Materials Science. Mater. Today Nano 2020, 11, 100087.
- Brahimi, M.; Boukhalfa, K.; Moussaoui, A. Deep Learning for Tomato Diseases: Classification and Symptoms Visualization. Appl. Artif. Intell. 2017, 31, 1–17.
- Arsenovic, M.; Karanovic, M.; Sladojevic, S.; Anderla, A.; Stefanovic, D. Solving Current Limitations of Deep Learning Based Approaches for Plant Disease Detection. Symmetry 2019, 11, 939.
- Peng, Y.; Wang, Y. An industrial-grade solution for agricultural image classification tasks. Comput. Electron. Agric. 2021, 187, 106253.
- Wang, Y.; Wang, J.; Zhang, W.; Zhan, Y.; Guo, S.; Zheng, Q.; Wang, X. A survey on deploying mobile deep learning applications: A systemic and technical perspective. Digit. Commun. Netw. 2021.
- Gao, J.; Westergaard, J.C.; Sundmark, E.H.R.; Bagge, M.; Liljeroth, E.; Alexandersson, E. Automatic late blight lesion recognition and severity quantification based on field imagery of diverse potato genotypes by deep learning. Knowl. Based Syst. 2021, 214, 106723.
More
Information
Subjects:
Agriculture, Dairy & Animal Science
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.8K
Revisions:
2 times
(View History)
Update Date:
12 Aug 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





