
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jianjun Qiao | + 1951 word(s) | 1951 | 2021-07-21 12:33:43 | | | |
| 2 | Ron Wang | + 167 word(s) | 2118 | 2021-08-10 08:26:59 | | |
Video Upload Options
Sesquiterpenes are important defense and signal molecules for plants to adapt to the environment, cope with stress, and communicate with the outside world, and their evolutionary history is closely related to physiological functions. In this study, the information of plant sesquiterpene synthases (STSs) with identified functions were collected and sorted to form a dataset containing about 500 members. The phylogeny of spermatophyte functional STSs was constructed based on the structural comparative analysis to reveal the sequence–structure–function relationships. We propose the evolutionary history of plant sesquiterpene skeletons, from chain structure to small rings, followed by large rings for the first time and put forward a more detailed function-driven hypothesis. Then, the evolutionary origins and history of spermatophyte STSs are also discussed. In addition, three newly identified STSs CaSTS2, CaSTS3, and CaSTS4 were analyzed in this functional evolutionary system, and their germacrene D products were consistent with the functional prediction. This demonstrates an application of the structure-based phylogeny in predicting STS function.
1. Introduction
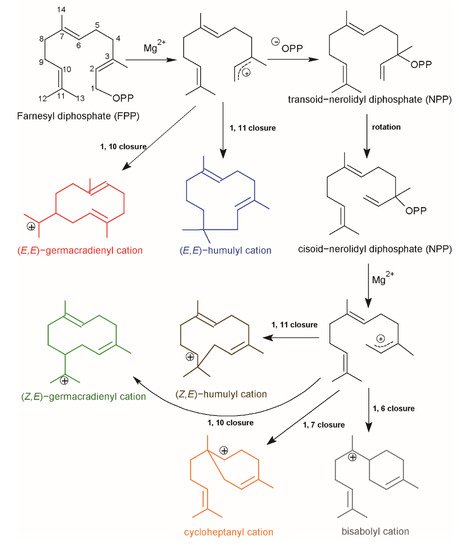
2. Current Insights
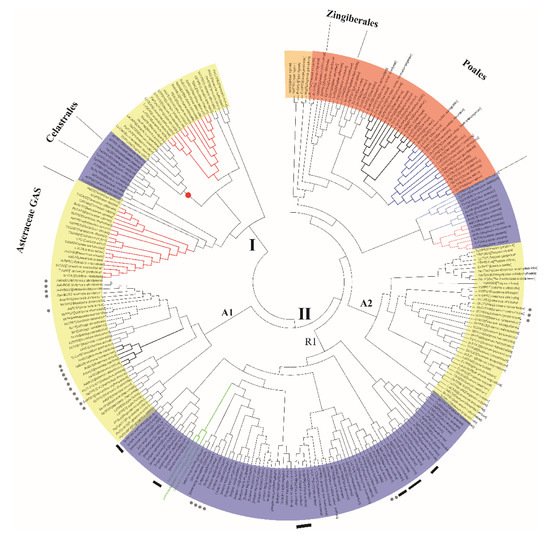
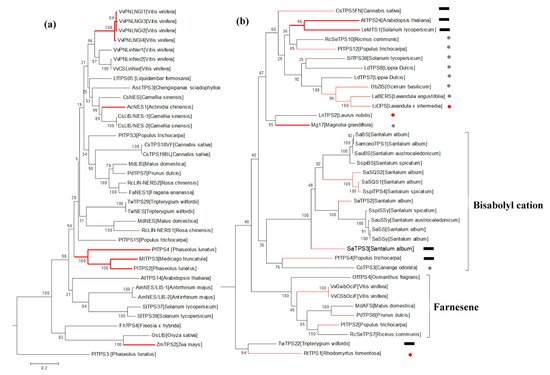
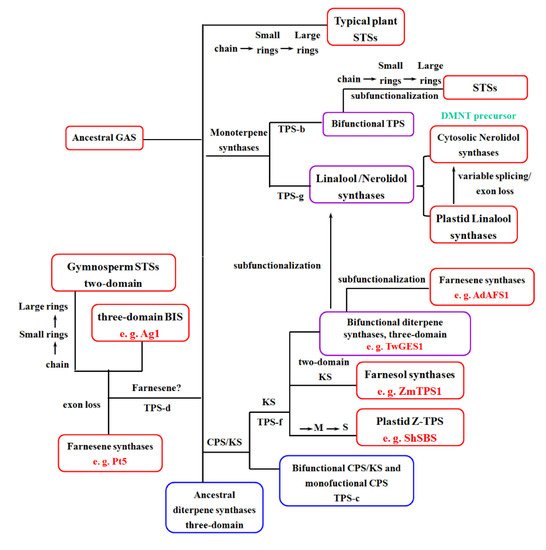
References
- Christianson, D.W. Structural and Chemical Biology of Terpenoid Cyclases. Chem. Rev. 2017, 117, 11570–11648.
- Fraga, B.M. Natural sesquiterpenoids. Nat. Prod. Rep. 2013, 30, 1226–1264.
- Gershenzon, J.; Dudareva, N. The function of terpene natural products in the natural world. Nat. Chem. Biol. 2007, 3, 408–414.
- Rasmann, S.; Köllner, T.G.; Degenhardt, J.; Hiltpold, I.; Toepfer, S.; Kuhlmann, U.; Gershenzon, J.; Turlings, T.C.J. Recruitment of entomopathogenic nematodes by insect-damaged maize roots. Nature 2005, 434, 732–737.
- Dreher, D.; Baldermann, S.; Schreiner, M.; Hause, B. An arbuscular mycorrhizal fungus and a root pathogen induce different volatiles emitted by Medicago truncatula roots. J. Adv. Res. 2019, 19, 85–90.
- Agrawal, A.A.; Heil, M. Synthesizing specificity: Multiple approaches to understanding the attack and defense of plants. Trends Plant Sci. 2012, 17, 239–242.
- Erb, M. Volatiles as inducers and suppressors of plant defense and immunity-origins, specificity, perception and signaling. Curr. Opin. Plant Biol. 2018, 44, 117–121.
- Tholl, D.; Lee, S. Terpene Specialized Metabolism in Arabidopsis thaliana. Arab. Book 2011, 9, e0143.
- Degenhardt, J.; Kllner, T.G.; Gershenzon, J. Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 2009, 69, 1621–1637.
- Davis, E.M.; Croteau, R. Cyclization enzymes in the biosynthesis of monoterpenes, sesquiterpenes, and diterpenes. In Biosynthesis: Aromatic Polyketides, Isoprenoids, Alkaloids; Leeper, F.J., Vederas, J.C., Eds.; Topics in Current Chemistry; Springer: Berlin/Heidelberg, Germany, 2000; Volume 209, pp. 53–95.
- Cao, R.; Zhang, Y.; Mann, F.M.; Huang, C.; Mukkamala, D.; Hudock, M.P.; Mead, M.E.; Prisic, S.; Wang, K.; Lin, F.-Y.; et al. Diterpene cyclases and the nature of the isoprene fold. Proteins-Struct. Funct. Bioinform. 2010, 78, 2417–2432.
- Gao, Y.; Honzatko, R.B.; Peters, R.J. Terpenoid synthase structures: A so far incomplete view of complex catalysis. Nat. Prod. Rep. 2012, 29, 1153–1175.
- Dudareva, N.; Martin, D.; Kish, C.M.; Kolosova, N.; Gorenstein, N.; Faldt, J.; Miller, B.; Bohlmann, J. (E)-beta-ocimene and myrcene synthase genes of floral scent biosynthesis in snapdragon: Function and expression of three terpene synthase genes of a new terpene synthase subfamily. Plant Cell 2003, 15, 1227–1241.
- Martin, D.M.; Faldt, J.; Bohlmann, J. Functional characterization of nine Norway spruce TPS genes and evolution of gymnosperm terpene synthases of the TPS-d subfamily. Plant Physiol. 2004, 135, 1908–1927.
- Durairaj, J.; Di Girolamo, A.; Bouwmeester, H.J.; de Ridder, D.; Beekwilder, J.; van Dijk, A.D.J. An analysis of characterized plant sesquiterpene synthases. Phytochemistry 2019, 158, 157–165.
- Pichersky, E.; Raguso, R.A. Why do plants produce so many terpenoid compounds? New Phytol. 2018, 220, 692–702.
- Richards, L.A.; Dyer, L.A.; Forister, M.L.; Smilanich, A.M.; Dodson, C.D.; Leonard, M.D.; Jeffrey, C.S. Phytochemical diversity drives plant-insect community diversity. Proc. Natl. Acad. Sci. USA 2015, 112, 10973–10978.
- Firn, R.D.; Jones, C.G. Natural products—A simple model to explain chemical diversity. Nat. Prod. Rep. 2003, 20, 382–391.
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011, 66, 212–229.
- Martin, D.M.; Bohlmann, J. Identification of Vitis vinifera (-)-alpha-terpineol synthase by in silico screening of full-length cDNA ESTs and functional characterization of recombinant terpene synthase. Phytochemistry 2004, 65, 1223–1229.
- Martin, D.M.; Aubourg, S.; Schouwey, M.B.; Daviet, L.; Schalk, M.; Toub, O.; Lund, S.T.; Bohlmann, J. Functional Annotation, Genome Organization and Phylogeny of the Grapevine (Vitis vinifera) Terpene Synthase Gene Family Based on Genome Assembly, FLcDNA Cloning, and Enzyme Assays. BMC Plant Biol. 2010, 10.
- Ma, L.-T.; Lee, Y.-R.; Liu, P.-L.; Cheng, Y.-T.; Shiu, T.-F.; Tsao, N.-W.; Wang, S.-Y.; Chu, F.-H. Phylogenetically distant group of terpene synthases participates in cadinene and cedrane-type sesquiterpenes accumulation in Taiwania cryptomerioides. Plant Sci. 2019, 289.
- Hillwig, M.L.; Xu, M.; Toyomasu, T.; Tiernan, M.S.; Wei, G.; Cui, G.; Huang, L.; Peters, R.J. Domain loss has independently occurred multiple times in plant terpene synthase evolution. Plant J. 2011, 68, 1051–1060.
- Trapp, S.C.; Croteau, R.B. Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics 2001, 158, 811–832.
- Loizzi, M.; Gonzalez, V.; Miller, D.J.; Allemann, R.K. Nucleophilic Water Capture or Proton Loss: Single Amino Acid Switch Converts -Cadinene Synthase into Germacradien-4-ol Synthase. Chembiochem 2018, 19, 100–105.
- Pazouki, L.; Niinemets, U. Multi-Substrate Terpene Synthases: Their Occurrence and Physiological Significance. Front. Plant Sci. 2016, 7.
- Gutensohn, M.; Orlova, I.; Nguyen, T.T.H.; Davidovich-Rikanati, R.; Ferruzzi, M.G.; Sitrit, Y.; Lewinsohn, E.; Pichersky, E.; Dudareva, N. Cytosolic monoterpene biosynthesis is supported by plastid-generated geranyl diphosphate substrate in transgenic tomato fruits. Plant J. 2013, 75, 351–363.
- Dong, L.; Jongedijk, E.; Bouwmeester, H.; Van Der Krol, A. Monoterpene biosynthesis potential of plant subcellular compartments. New Phytol. 2016, 209, 679–690.
- McAndrew, R.P.; Peralta-Yahya, P.P.; DeGiovanni, A.; Pereira, J.H.; Hadi, M.Z.; Keesling, J.D.; Adams, P.D. Structure of a Three-Domain Sesquiterpene Synthase: A Prospective Target for Advanced Biofuels Production. Structure 2011, 19, 1876–1884.
- Luck, K.; Chen, X.; Norris, A.M.; Chen, F.; Gershenzon, J.; Koellner, T.G. The reconstruction and biochemical characterization of ancestral genes furnish insights into the evolution of terpene synthase function in the Poaceae. Plant Mol. Biol. 2020, 104, 203–215.
- Chang, Y.J.; Song, S.H.; Park, S.H.; Kim, S.U. Amorpha-4,11-diene synthase of Artemisia annua: cDNA isolation and bacterial expression of a terpene synthase involved in artemisinin biosynthesis. Arch. Biochem. Biophys. 2000, 383, 178–184.
- Muangphrom, P.; Seki, H.; Suzuki, M.; Komori, A.; Nishiwaki, M.; Mikawa, R.; Fukushima, E.O.; Muranaka, T. Functional Analysis of Amorpha-4,11-Diene Synthase (ADS) Homologs from Non-Artemisinin-Producing Artemisia Species: The Discovery of Novel Koidzumiol and (+)-alpha-Bisabolol Synthases. Plant Cell Physiol. 2016, 57, 1678–1688.
- Tantillo, D.J. Biosynthesis via carbocations: Theoretical studies on terpene formation. Nat. Prod. Rep. 2011, 28, 1035–1053.
- Blank, P.N.; Shinsky, S.A.; Christianson, D.W. Structure of Sesquisabinene Synthase 1, a Terpenoid Cyclase That Generates a Strained 3.1.0 Bridged-Bicyclic Product. Acs Chem. Biol. 2019, 14, 1011–1019.
- Koo, H.J.; Vickery, C.R.; Xu, Y.; Louie, G.V.; O’Maille, P.E.; Bowman, M.; Nartey, C.M.; Burkart, M.D.; Noel, J.P. Biosynthetic potential of sesquiterpene synthases: Product profiles of Egyptian Henbane premnaspirodiene synthase and related mutants. J. Antibiot. 2016, 69, 524–533.
- Gennadios, H.A.; Gonzalez, V.; Di Costanzo, L.; Li, A.; Yu, F.; Miller, D.J.; Allemann, R.K.; Christianson, D.W. Crystal Structure of (+)-delta-Cadinene Synthase from Gossypium arboreum and Evolutionary Divergence of Metal Binding Motifs for Catalysis. Biochemistry 2009, 48, 6175–6183.
- Hsieh, F.-L.; Chang, T.-H.; Ko, T.-P.; Wang, A.H.J. Structure and Mechanism of an Arabidopsis Medium/Long-Chain-Length Prenyl Pyrophosphate Synthase. Plant Physiol. 2011, 155, 1079–1090.
- Jia, Q.; Li, G.; Kollner, T.G.; Fu, J.; Chen, X.; Xiong, W.; Crandall-Stotler, B.J.; Bowman, J.L.; Weston, D.J.; Zhang, Y.; et al. Microbial-type terpene synthase genes occur widely in nonseed land plants, but not in seed plants. Proc. Natl. Acad. Sci. USA 2016, 113, 12328–12333.
- Suetsugu, K.; Kawakita, A.; Kato, M. Host range and selectivity of the hemiparasitic plant Thesium chinense (Santalaceae). Ann. Bot. 2008, 102, 49–55.
- Rice, D.W.; Alverson, A.J.; Richardson, A.O.; Young, G.J.; Virginia Sanchez-Puerta, M.; Munzinger, J.; Barry, K.; Boore, J.L.; Zhang, Y.; dePamphilis, C.W.; et al. Horizontal Transfer of Entire Genomes via Mitochondrial Fusion in the Angiosperm Amborella. Science 2013, 342, 1468–1473.
- Barkman, T.J.; McNeal, J.R.; Lim, S.-H.; Coat, G.; Croom, H.B.; Young, N.D.; dePamphilis, C.W. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. Bmc Evol. Biol. 2007, 7.





