Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Tina Kabelitz | + 3733 word(s) | 3733 | 2021-07-20 04:42:14 | | | |
| 2 | Vivi Li | Meta information modification | 3733 | 2021-08-05 03:17:46 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Kabelitz, T.; Fulde, M. Streptococcus spp. in Bovine Mastitis. Encyclopedia. Available online: https://encyclopedia.pub/entry/12750 (accessed on 07 February 2026).
Kabelitz T, Fulde M. Streptococcus spp. in Bovine Mastitis. Encyclopedia. Available at: https://encyclopedia.pub/entry/12750. Accessed February 07, 2026.
Kabelitz, Tina, Marcus Fulde. "Streptococcus spp. in Bovine Mastitis" Encyclopedia, https://encyclopedia.pub/entry/12750 (accessed February 07, 2026).
Kabelitz, T., & Fulde, M. (2021, August 04). Streptococcus spp. in Bovine Mastitis. In Encyclopedia. https://encyclopedia.pub/entry/12750
Kabelitz, Tina and Marcus Fulde. "Streptococcus spp. in Bovine Mastitis." Encyclopedia. Web. 04 August, 2021.
Copy Citation
The Streptococcus genus belongs to one of the major pathogen groups inducing bovine mastitis. In the dairy industry, mastitis is the most common and costly disease. It not only negatively impacts economic profit due to milk losses and therapy costs, but it is an important animal health and welfare issue as well.
dairy
cow
mastitis
bovine
streptococci
udder
infection
inflammation
livestock
milk
1. Introduction
Over 130 pathogens are known to be associated with bovine mastitis [1][2], some of them belonging to the Streptococcus genus. Streptococci are gram-positive bacteria of spherical shape (0.5–2 µm) that usually form pairs or chains. They are classified on the basis of colony morphology, hemolysis, and serologic specificity into the Lancefield group taxonomic system. Many of them are facultative anaerobe, non-pathogenic and belong to the commensal microbiota of humans and animals. However, some streptococci can cause severe diseases and health issues, such as bovine mastitis. Here, the most relevant species are S. agalactiae, S. dysgalactiae ssp. dysgalactiae (hereinafter referred to as S. dysgalactiae) and S. uberis. Streptococcal pathogens rarely associated with bovine mastitis are S. canis, S. lutetiensis and S. equinus.
Mastitis is the most common and costly disease in dairy industry and of worldwide relevance [3][4]. The economic losses of mastitis are calculated with 124€ (=147$) per cow per year, resulting in losses of 500 million, 3 and 125 billion € in Germany, the EU and worldwide, respectively [5]. Global bovine mastitis rates are typically between 30–50% of all cows per year [3][6]. Next to the financial losses due to less milk yield and quality, the veterinary treatment, medication, and increased personnel expenses, mastitis is an important issue of animal welfare and the main reason for dairy cow culling. Mastitis infected cows can show a wide range of symptoms: swelling, heat and pain of the udder, milk with abnormal appearance, increased body temperature, lethargy, and anorexia [6][7]. Bovine mastitis can be classified into three classes according to the inflammation degree: clinical, subclinical, and chronic [5]. Clinical mastitis is characterized by visible abnormalities of cow and milk, which is not the case for subclinical mastitis. Here, only the milk yield and somatic cell count in the milk are changed. The incidence of subclinical mastitis is estimated to be 15–40 times higher than for clinical mastitis [8]. Therefore, subclinical mastitis is economically more relevant due to its higher frequency and capacity to reduce milk yields while going unnoticed. The average duration of a streptococcal mastitis is 12 days but can be prolonged for >300 days in chronic cases [9]. If an acute mastitis is not successfully cured, it can become chronic and lead to reduced fertility [4]. S. uberis and S. agalactiae are well-known pathogens able to induce chronic mastitis [10][11]. Treatment and prophylaxis of mastitis are the most common reasons for antibiotic usage in dairy cows [12][13], bearing the risk of enhanced selection in favor of antimicrobial resistant microorganisms [1][5].
2. Classification
Streptococci are reported to be among the main pathogens causing bovine mastitis all over the world [9][14]. Mastitis pathogens can be classified in contagious and environmental [5]. Contagious pathogens are adapted to survive within the host and they spread from cow to cow primarily through the milking process. Contagious bacteria have the potential to spread within a herd easily and widely. In contrast, environmental pathogens are able to survive outside the host and are part of the normal microflora of the cow’s vicinity. Exposure through environmental streptococci occurs during and between milking, during the dry period or prior parturition of heifers [9]. The pathogen exposure is related to their environmental abundance, which is influenced, e.g., by humidity and temperature. Environmental pathogens invade the udder when the teat channel is opened after milking or after damage.
S. uberis is primarily environmental, however cases of contagion have been observed [15]. The species is mostly alpha-hemolytic, capable of partial hemolysis, but has also been shown to be non-hemolytic in some cases. Biochemical identification is facilitated by a variable CAMP (Christine–Atkinson–Munch–Peterson test) phenotype as well as aesculin, sodium hippurate and inulin degradation. Global Lancefield classification of S. uberis is quite challenging, since some strains have been shown to be Lancefield E, G, P or U positive. Since the first isolation of S. uberis from a bovine mastitis case in 1932, the pathogen has been detected in a variety of bovine host infections such as lactating cows, dry cows, heifers, and multiparous cows [15].
For S. dysgalactiae, the classification in environmental or contagious is not clear. Due to its ability to survive within the host and in the environment, it is described as an intermediate pathogen [16]. The majority of S. dysgalactiae strains are non-hemolytic, although alpha-hemolytic exceptions exist. Phenotypically, it is CAMP negative, does not degrade aesculin and is usually classified Lancefield group C. S. dysgalactiae is primarily associated with bovine infections, but other ruminants, such as goats or sheep, may be affected as well.
S. agalactiae is a contagious pathogen but may also colonize the gastrointestinal tract of dairy cows. In bovine mammary glands, S. agalactiae can survive indefinitely by forming biofilms and is heavily associated with subclinical mastitis [5]. S. agalactiae is within the Lancefield group B classification. The bacterium is generally considered beta-hemolytic, but some non-hemolytic strains have been observed and CAMP positive. S. agalactiae has nine distinctive serotypes labeled Ia, Ib, II, III, IV, V, VI, VII, and VIII, with a tenth serotype labeled IX discovered in 2007. Pathogenicity of S. agalactiae varies with its serotype. Next to dairy, this species is a highly relevant human pathogen at early ages, since it is among the most common causes of bacterial meningitis in neonates. In the United States, between 2005 and 2006, the most common S. agalactiae serotype implicated in invasive human diseases was serotype V, accounting for more than 29% of the recorded cases at the time, followed by serotypes Ia, II and III [17]. A myriad of cases have been shown that S. agalactiae can be hosted by piscine and aquatic mammals as well. Multilocus sequence typing has allowed the description of the phylogenetic relations between the different hosts of S. agalactiae by assigning sequence types (ST) based on their allele numbers. Some strains from piscine hosts were shown to be pathogenic to humans as well. Interestingly, most human STs are quite distinct from bovine STs, apart from the bovine ST-23 and ST-61 strains, which have a genetic relation with human ST-23 and ST-17. These two strains belong to serotypes Ia and III, respectively, and are heavily associated with neonatal infections [18][19][20].
S. canis belongs to the contagious pathogens and is grouped to Lancefield group G. It shows a beta-hemolytic, CAMP negative, and aesculin hydrolyzing phenotype. S. equinus is classified as environmental pathogen and to the Lancefield group D. Its phenotype is a variable hemolysis, CAMP negative and aesculin hydrolysis positive. S. lutetiensis is a contagious streptococcal species within the Lancefield D group. It is characterized by alpha-hemolysis and a CAMP negative, aesculin hydrolyzing phenotype.
3. Pathogenesis and Virulence Factors
During the infectious process, streptococci face a diversity of changing environments and hence need to adapt to varying conditions for successful infection. As a consequence, they express a number of virulence factors that allow survival and replication in different host tissues during pathogenesis, e.g., by conferring adhesion and invasion to host cells or avoidance of immune recognition and subsequent degradation. The virulence factors of S. uberis, S. dysgalactiae, and S. agalactiae are summarized in Figure 2.
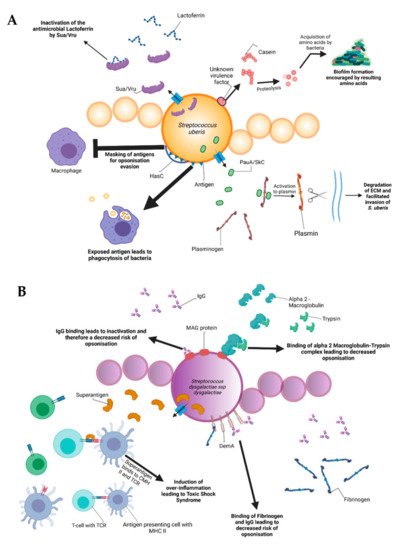
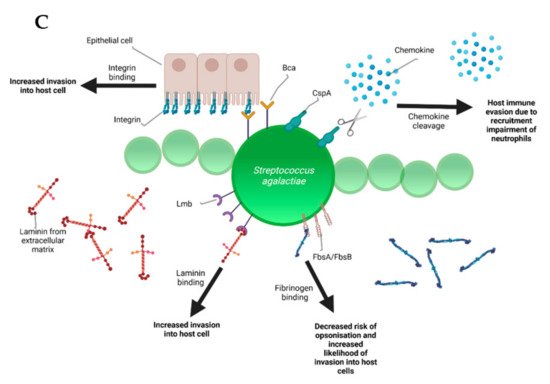
Figure 2. Schematic representation of the bovine-associated streptococcal virulence factors. (A) S. uberis harbors the capacity for preventing opsonization via the synthesis of the protein HasC, capable of masking the antigens present on the surface of S. uberis. Synthesis of Sua and Vru proteins allow binding of the antimicrobial lactoferrin and thus creating a less hostile environment for the pathogen. Lysis of casein through a so far unknown process provides extra nutrients and thereby favors biofilm formation. PauA and SkC proteins confer plasminogen-activating capabilities to S. uberis. The activation of plasminogen to plasmin leads to the degradation of the extracellular matrix, which in turn facilitates bovine mammary epithelial cell invasion. (B) S. dysgalactiae is equally capable of avoiding opsonization via the binding of fibrinogen with the M-like protein DemA, the binding of IgG and the binding of the alpha 2 macroglobulin-trypsin complex via the synthesis of the MAG protein. (C) S. agalactiae possesses virulence factors implicated in invasion of host cells, namely Bca for integrin binding and Lmb for laminin binding. Immune evasion is facilitated via the synthesis of proteins such as FbsA and FbsB implicated in fibrinogen binding. This leads to a decreased risk of opsonization by phagocytic cells. Synthesis of the serine protease CspA allows S. agalactiae to cleave certain chemokines, which are responsible for neutrophil recruitment, further increasing immune evasion potential.
Amongst all the listed virulence factors, a study of 78 S. uberis strains showed that the three most prevalent are the hasC gene (hyaluronic acid production) present in 89.7% of the studied strains, followed by the sua gene (lactoferrin binding) at 83.3% and gapC (plasmin binding as well as immunomodulation) at 79.4%. The has operon is a conserved gene region present in all strains of group A streptococci and encapsulated group C streptococci. It enables production of a hyaluronic acid capsule that protects e.g., S. uberis from opsonization and phagocytosis and mediates resistance to bacterial clearance within neutrophil extracellular traps [21]. In terms of capsulation, the most well-characterized virulence factors are the capsular polysaccharides coded by the cps and neu genes, which are at the basis of determining different Group-B-Streptococci (GBS) serotypes discussed later. The capsular types are labeled as Ia, Ib, II-IX and are critical for grouping the different pathogenic types of GBS. These proteins confer immune evasion by masking pro-inflammatory cell wall components and by mimicry of host-cell surface glycoconjugates. SodA and rhamnopolyene also contribute to immune evasion by detoxifying superoxide responsible for the formation of reactive oxygen species implicated in oxidative stress. The C5a peptidase coded by scpB is another virulence factor that contributes to immune evasion. C5a is a component of the human complement system, its degradation leads to an inhibited opsonophagocytic killing pathway. In addition to this, the peptidase has been shown to bind to fibronectin, thus contributing to the bacteria’s adhesion and invasion in epithelial cells [22].
Internalization of S. uberis into mammary epithelial cells is an important early event in the establishment of mastitis in dairy cows. Possession of the S. uberis Adhesion Molecule (SUAM), which was suspected to have an affinity to the host protein lactoferrin, seems to facilitate pathogenesis in this context. Lactoferrin is a protein present in human and bovine milk as well as many other mammalian body fluids. The protein has antimicrobial properties as it binds iron, which is essential for bacterial and viral replication [23]. Since iron demand of streptococcal species is rather low, lactoferrin was demonstrated to have a bacteriostatic effect on different streptococci [24]. Clinical trials attempted on seven Holstein Friesian cows showed that 67% of the mammary quarters infected with S. uberis wild-type showed clinical manifestations of mastitis (clots in produced milk, firm and moderate swelling of the mammary glands with red coloration). In contrast, experiments conducted with a SUAM-deficient mutant only manifested mastitis in 28.6% of the infected quarters. Colony counts in milk samples as well as somatic cell counts displayed decreased S. uberis abundance in the absence of SUAM compared to the wild-type, indicating that the sua gene could be an important virulence factor. Moreover, SUAM facilitated efficient epithelial cell adhesion and subsequent internalization [25]. However, other studies have shown that certain factors have overlapping functions with sua in terms of pathogenesis of S. uberis. Similarly to sua, vru gene deletion has been shown to decrease the pathogenicity of S. uberis, probably due to its capability of binding the antimicrobial lactoferrin [26]. This is an indicator that the sua gene should not be considered the only virulence factor of S. uberis and that other genes should be taken into account for further research.
Lactoferrin plays a role in pathogenesis of other streptococci as well. After 8 h of exposure to lactoferrin, a dose-dependent growth inhibition of S. dysgalactiae was observed in a previous study due to the antimicrobial properties of the protein. In addition, assays conducted with HC-11 murine epithelial cells revealed lactoferrin-dependent decrease of bacterial adhesion [24]. Lactoferrin may serve as a bridging molecule for S. uberis, consequently promoting adherence and internalization into mammary epithelial cells, thereby surviving host defense mechanisms by immune evasion [27]. Although S. dysgalactiae also expresses lactoferrin-binding proteins, allowing interaction [28] similar to S. uberis, the effect of lactoferrin on S. dysgalactiae pathogenesis is quite opposite. The mechanisms of internalization and host cell interaction after lactoferrin binding of S. dysgalactiae are poorly defined and require further classification to explain these contrary phenotypes.
Besides lactoferrin-binding proteins, adherence and invasion are also extensively studied for S. agalactiae. Exposure of bovine mammary epithelial cells to S. agalactiae significantly decreased host cell viability starting 2 h post infection. Electron microscopy showed that adherence of the bacteria was achieved 6 h post-infection with cell membrane breakage visible after 8 h [29]. Furthermore, host-cell adherence and invasion are facilitated by extracellular matrix protein binding. The main virulence factors for this are the fibrinogen binding proteins FbsA and FbsB, which promote entry into the host cell and by the laminin binding protein Lmb. Serine-rich repeat proteins allow for S. agalactiae to adhere to human keratin and epithelial cells. The alpha C protein coded by bca is also shown to participate in cell adherence to epithelial cells [22]. S. dysgalactiae was found to be capable of binding extracellular matrix proteins such as fibronectin, a main component of connective tissues. This could encourage bacterial adherence and internalization, e.g., to the mammary tissue as can be observed in bovine mastitis [30]. Furthermore, binding of S. dysgalactiae to vitronectin was implicated in phagocytosis. Exposure of S. dysgalactiae to vitronectin before phagocytic assays increased phagocytosis by bovine polymorphonuclear neutrophils [31].
Binding to extracellular matrix proteins is also a key aspect of biofilm formation, which comprises one of the main pathogenic factors of many bacteria and streptococci are no exception. It was previously demonstrated that the addition of milk to liquid THY (Todd–Hewitt broth with 1%, w/v yeast extract) culture led to an encouragement of biofilm formation by up to 800%. A more detailed look revealed that alpha and beta casein were at least partially responsible for the phenomenon that was decreased by the addition of protease inhibitors. It was speculated that the casein proteins were most likely degraded by proteases, thereby allowing S. uberis to utilize the resulting amino acids as nutrient sources [32]. Further studies on S. uberis biofilm formation showed that certain genes associated with biofilm formation were more prevalent than others. luxS and comEA (corresponding to quorum sensing and bacterial competence, respectively) were found at rates of 42.8% and 21.4% of the S. uberis isolates which were used for the study. Given the pathogenic capabilities of biofilms, it would be interesting to unravel underlying mechanisms in order to diminish invasive capabilities of S. uberis [33]. Furthermore, biofilm formation is suspected to play an important role in S. dysgalactiae pathogenesis in cases of bovine mastitis as well. An article by Alves-Barroco and colleagues in 2019 showed that certain strains of S. dysgalactiae were capable of forming biofilms on hydrophilic surfaces and that treatment of these biofilms with the protein fisetin induced a decrease in biofilm formation. Fisetin is a flavonol that exhibits various effects, including neurotrophic, antioxidant, anti-inflammatory, and anti-angiogenic effects [34]. Furthermore, a brpA-like gene might contribute to the initial steps of biofilm formation. A recent study postulates that this brpA-like protein is inhibited by fisetin, thus rendering the gene a potential target to eliminate S. dysgalactiae biofilm formation [35].
Plasminogen activation is yet another virulence mechanism prevalent in the streptococcal genus. Secreted polypeptides, such as streptokinase of S. dysgalactiae, can degrade fibrin and connective tissue, thereby allowing deeper tissue infiltration. After exposure to streptokinase, plasminogen is thereby activated to plasmin, in turn allowing the hydrolysis of connective tissue proteins and subsequent tissue penetration [36]. Besides plasminogen degradation, e.g., by streptokinase, S. dysgalactiae produces a hyaluronidase, which degrades hyaluronic acid. Hyaluron is a polysaccharide present in connective tissues and its degradation is thought to contribute to the tissue invasive properties of streptococci. The hly gene found in S. agalactiae is a virulence factor that codes for hyaluronate lyase. The protein is capable of cleaving hyaluronan, allowing for an easier invasion and spread of GBS, similar to the action of streptokinase in S. dysgalactiae [37][38]. pauA is notable since it was the first described plasminogen activator affecting bovine plasminogen. Furthermore, it encourages the hydrolysis of casein to peptides allowing the bacteria to profit from the resulting amino acids [33]. skC is another gene which codes for a plasminogen activator. A study by Loures in Brazil has shown that this gene, along with pauA, is highly conserved between different strains of S. uberis [39].
S. dysgalactiae has a vast array of potential virulence factors, e.g., facilitating binding to diverse host structures that could push towards potential vaccine agents. For instance, S. dysgalactiae is capable of binding IgG, therefore interfering with immune responses such as opsonization and toxin neutralization [40]. In addition to IgG binding, a protein named MAG for alpha2-Macroglobulin/Albumin/IgG-binding can additionally connect to alpha2-macroglobulin and is suspected to contribute to the inhibition of opsonization as described previously [41]. It was shown that when alpha2M was associated with trypsin to form alpha2M-T, S. dysgalactiae can attach to the complex, thus creating a dose-dependent inhibition of phagocytosis, thereby significantly contributing to pathogenesis [42].
Another notable virulence factor for immune evasion is the serine protease coded by cspA. This protease cleaves fibrinogen and chemokines, but also impairs neutrophil recruitment and phagocytic killing. Serotype III of GBS has been shown to be capable of producing a protein called Rib coded by the rib gene. Rib is suspected to be one of the reasons for S. agalactiae host-immune evasion thanks to a structure that confers domain atrophy (a phenomenon where protein domains lose a significant number of core structural elements) [43]. One final important virulence factor in terms of immune evasion is a superantigen named S. dysgalactiae-derived mitogen. Superantigens are a class of antigens capable of inciting an excessive activation of the immune system, thereby inducing potentially life-threatening symptoms such as shock [36].
A versatile variety of other virulence factors facilitate additional essential steps in streptococcal pathogenesis. In terms of bovine mastitis, survival in milk is clearly an important step besides invasion of mammary epithelial cells. The pathogenicity of S. agalactiae varies and certain strains are more adapted to bovine hosts in that they are more capable of growing in cow milk than their human or fish-associated counterparts. An article by Maoda Pang and others observed that bovine strains achieved a CFU/mL count of around 5 × 109 as opposed to the 108 CFU/mL of the human and fish strains investigated in this study after 12 h of growth in milk. Furthermore, biofilm formation was more important in bovine-associated strains, showing ODs that doubled or even tripled the ones evaluated in the other strains [44]. SCC indicates the total number of cells per milliliter in milk, which is increased during infection due to the resulting inflammation. According to the 2013 Bulletin of the International Dairy Federation, milk is considered abnormal when SCCs are higher than 200,000 cells/mL [45]. SCCs, determined with an automated, fluorescent, microscopic somatic cell counter, were increased when milk samples contained S. dysgalactiae. Indirect fluorescence labeling targeting neutrophils showed that there was a particular increase in neutrophil recruitment [46]. In addition, a notable increase in Il-1beta and TNF-alpha expression, both being potent inducers of the acute immune response, has been observed via real-time PCR. Further understanding these phenomena could push towards a deeper comprehension of S. dysgalactiae pathogenesis [47]. Different virulence traits, however, enable survival of streptococci besides inflammatory responses of the host. For instance, extracellular deoxyribonucleases are shown to aid in bacterial proliferation thanks to the neutrophil-mediated resistance it confers. The enzyme is capable of degrading the antimicrobial system induced by the neutrophils. It was shown in a study by Florindo that all of the S. agalactiae strains associated with subclinical or clinical mastitis in 121 clinical samples were capable of synthesizing DNAses [48]. This could imply that DNAse synthesis could be important for the development of bovine mastitis. Further research is required.
M-proteins are the most important virulence factors in streptococcal pathogenesis. It is extensively described in S. pyogenes pathogenesis although it is a virulence factor present in a wide variety of streptococcal species. Its best-known property is the capacity to inhibit phagocytosis in non-immune humans. However, it has been associated with a variety of functions and was shown to interact with an extensive amount of host proteins, such as fibrinogen, albumin, plasminogen, and immunoglobulins. The N-terminal part of the protein is hyper-variable, leading to antigenic diversity, consequently allowing efficient serotyping of S. pyogenes. The N-terminal region is also known to induce protective antibody production, turning it into a promising vaccine candidate. In contrast, the C-terminal region is highly conserved among M proteins and can bind to a wide array of host molecules [49]. An M-like protein was discovered in S. dysgalactiae and S. canis, termed DemA and SCM (S. canis M-protein), respectively. It displays plasma protein binding properties and sequence similarities with the M-protein of S. pyogenes. Given the importance of the protein in Group A Streptococcus, it would be interesting to further study the effect of DemA in S. dysgalactiae pathogenesis [50].
Group B Streptococcus has a huge set of virulence factors that are quite prevalent in most strains isolated from humans. They have been shown to have a wide variety of functions, such as pore-formation via toxins, immune evasion, resistance to antimicrobial peptides, host-cell adherence and invasion. Pore formation via toxins is another main virulence mechanism of S. agalactiae. Beta-hemolysin or cytolysins are significant virulence factors, which promote invasion of host cells, but also impair cardiac and liver functions whilst inducing inflammatory responses and apoptosis. The CAMP factor is a pore-forming toxin, which attacks the host-cell membrane and binds to GPI anchored proteins. Resistance to antimicrobial peptides (AMPs) is an interesting virulence mechanism for S. agalactiae. Alanylation of lipotechoic acid decreases cell surface charge, thus repelling AMPs. In addition, S. agalactiae has been shown to bind penicillin via a protein labeled PBP1a through a so far unknown mechanism. The expression of pili also contributes to AMP resistance, although not much is known about this process either.
Bovine strains possess much of the same virulence factors as the human strains, yet to varying degrees. In an article by Mohammad Emaneini in 2016, it was shown that 89% of the 48 bovine strains expressed the rib gene, a value significantly higher than that observed in human isolates, indicating that this gene could be an important virulence factor for bovine-specific cases. However, all of the other virulence factors typically associated with human infections were not detected in bovine isolates. This is in direct contrast with a study by Duarte in 2005, where he showed that the majority of his 38 bovine isolates expressed scpB and bca. Even the findings concerning the rib gene by Emaneini are in direct contrast with a study by Jain in 2012. Jain showed that only 26% of bovine isolates possessed the gene, an incredibly low number, compared to the 89% observed by Emaneini. Overall, determining bovine streptococci mastitis specific virulence factors by comparing those found in human isolates seems to have variable results. The bacteria most likely have bovine-specific virulence factors that are fairly different from those found in human isolates [10][37][51].
Virulence factors for S. canis and S. equinus are rarely described. For S. lutetiensis known virulence factors are: hemolysin (Hly), α C protein (Bca), superantigen proteins, and C5a peptidase (ScpB) [52].
References
- Bradley, A.J. Bovine Mastitis: An Evolving Disease. Vet. J. 2002, 164, 116–128.
- Leigh, J. Streptococcus uberis: A permanent barrier to the control of bovine mastitis? Vet. J. 1999, 157, 225–238.
- Hogeveen, H.; Huijps, K.; Lam, T.J. Economic aspects of mastitis: New developments. N. Z. Vet. J. 2011, 59, 16–23.
- Ruegg, P.L. A 100-Year Review: Mastitis detection, management, and prevention. J. Dairy Sci. 2017, 100, 10381–10397.
- Cheng, W.N.; Han, S.G. Bovine mastitis: Risk factors, therapeutic strategies, and alternative treatments—A review. Asian Australas J. Anim. Sci. 2020, 33, 1699–1713.
- Heringstad, B.; Klemetsdal, G.; Ruane, J. Selection for mastitis resistance in dairy cattle: A review with focus on the situation in the Nordic countries. Livest. Prod. Sci. 2000, 64, 95–106.
- Kibebew, K. Bovine mastitis: A review of causes and epidemiological point of view. J. Biol. Agric. Healthc. 2017, 7, 1–14.
- Martin, P.; Barkema, H.; Brito, L.; Narayana, S.; Miglior, F. Symposium review: Novel strategies to genetically improve mastitis resistance in dairy cattle. J. Dairy Sci. 2018, 101, 2724–2736.
- Hogan, J.S.; Smith, K.L. Environmental streptococcal mastitis: Facts, fables, and fallacies. In Proceedings of the Fifth International Dairy Housing Conference, Fort Worth, TX, USA, 29–31 January 2003; pp. 9–16.
- Jain, B.; Tewari, A.; Bhandari, B.B.; Jhala, M.K. Antibiotic resistance and virulence genes in Streptococcus agalactiae isolated from cases of bovine subclinical mastitis. Vet. Arhiv 2012, 82, 423–432.
- Benić, M.; Maćešić, N.; Cvetnić, L.; Habrun, B.; Cvetnić, Ž.; Turk, R.; Đuričić, D.; Lojkić, M.; Dobranić, V.; Valpotić, H. Bovine mastitis: A persistent and evolving problem requiring novel approaches for its control-a review. Vet. Arhiv 2018, 88, 535–557.
- Botrel, M.-A.; Haenni, M.; Morignat, E.; Sulpice, P.; Madec, J.-Y.; Calavas, D. Distribution and Antimicrobial Resistance of Clinical and Subclinical Mastitis Pathogens in Dairy Cows in Rhône-Alpes, France. Foodborne Pathog. Dis. 2009, 7, 479–487.
- Gomes, F.; Henriques, M. Control of Bovine Mastitis: Old and Recent Therapeutic Approaches. Curr. Microbiol. 2016, 72, 377–382.
- Kaczorek, E.; Małaczewska, J.; Wójcik, R.; Rękawek, W.; Siwicki, A.K. Phenotypic and genotypic antimicrobial susceptibility pattern of Streptococcus spp. isolated from cases of clinical mastitis in dairy cattle in Poland. J. Dairy Sci. 2017, 100, 6442–6453.
- Oliver, S.P.; Pighetti, G.M. Mastitis Pathogens|Environmental Pathogens. Encycl. Dairy Sci. 2002, 1728–1734.
- Wente, N.; Krömker, V. Streptococcus dysgalactiae—Contagious or Environmental? Animals 2020, 10, 2185.
- Raabe, V.N.; Shane, A.L. Group B streptococcus (Streptococcus agalactiae). Gram Posit. Pathog. 2019, 228–238.
- Evans, J.J.; Bohnsack, J.F.; Klesius, P.H.; Whiting, A.A.; Garcia, J.C.; Shoemaker, C.A.; Takahashi, S. Phylogenetic relationships among Streptococcus agalactiae isolated from piscine, dolphin, bovine and human sources: A dolphin and piscine lineage associated with a fish epidemic in Kuwait is also associated with human neonatal infections in Japan. J. Med. Microbiol. 2008, 57, 1369–1376.
- Lyhs, U.; Kulkas, L.; Katholm, J.; Waller, K.P.; Saha, K.; Tomusk, R.J.; Zadoks, R.N. Streptococcus agalactiae serotype IV in humans and cattle, northern Europe. Emerg. Infect. Dis. 2016, 22, 2097.
- Pereira, U.; Mian, G.; Oliveira, I.; Benchetrit, L.; Costa, G.; Figueiredo, H. Genotyping of Streptococcus agalactiae strains isolated from fish, human and cattle and their virulence potential in Nile tilapia. Vet. Microbiol. 2010, 140, 186–192.
- Cole, J.N.; Pence, M.A.; Köckritz-Blickwede, M.V.; Hollands, A.; Gallo, R.L.; Walker, M.J.; Nizet, V.; Norrby-Teglund, A.; Low, D.E. M Protein and Hyaluronic Acid Capsule Are Essential for In Vivo Selection of covRS Mutations Characteristic of Invasive Serotype M1T1 Group A Streptococcus. Mbio 2010, 1, e00191-10.
- Rajagopal, L. Understanding the regulation of Group B Streptococcal virulence factors. Future Microbiol. 2009, 4, 2.
- Kell, D.B.; Heyden, E.L.; Pretorius, E. The biology of lactoferrin, an iron-binding protein that can help defend against viruses and bacteria. Front. Immunol. 2020, 11, 1221.
- O’Halloran, F.; Beecher, C.; Chaurin, V.; Sweeney, T.; Giblin, L. Lactoferrin affects the adherence and invasion of Streptococcus dysgalactiae ssp. dysgalactiae in mammary epithelial cells. J. Dairy Sci. 2016, 99, 4619–4628.
- Almeida, R.A.; Dego, O.K.; Headrick, S.I.; Lewis, M.J.; Oliver, S.P. Role of Streptococcus uberis adhesion molecule in the pathogenesis of Streptococcus uberis mastitis. Vet. Microbiol. 2015, 179, 332–335.
- Egan, S.A.; Ward, P.N.; Watson, M.; Field, T.R.; Leigh, J.A. Vru (Sub0144) controls expression of proven and putative virulence determinants and alters the ability of Streptococcus uberis to cause disease in dairy cattle. Microbiology 2012, 158, 1581–1592.
- Patel, D.; Almeida, R.A.; Dunlap, J.R.; Oliver, S.P. Bovine lactoferrin serves as a molecular bridge for internalization of Streptococcus uberis into bovine mammary epithelial cells. Vet. Microbiol. 2009, 137, 297–301.
- Park, H.-M.; Almeida, R.; Luther, D.; Oliver, S. Binding of bovine lactoferrin to Streptococcus dysgalactiae subsp. dysgalactiae isolated from cows with mastitis. FEMS Microbiol. Lett. 2002, 208, 35–39.
- Tong, J.; Sun, M.; Zhang, H.; Yang, D.; Zhang, Y.; Xiong, B.; Jiang, L. Proteomic analysis of bovine mammary epithelial cells after in vitro incubation with S. agalactiae: Potential biomarkers. Vet. Res. 2020, 51, 1–14.
- Mamo, W.; Fröman, G.; Sundås, A.; Wadström, T. Binding of fibronectin, fibrinogen and type II collagen to streptococci isolated from bovine mastitis. Microb. Pathog. 1987, 2, 417–424.
- Filippsen, L.F. Bovine S protein (vitronectin) increases phagocytosis of Streptococcus dysgalactiae. Rev. Microbiol. 1999, 30, 15–18.
- Varhimo, E.; Varmanen, P.; Fallarero, A.; Skogman, M.; Pyörälä, S.; Iivanainen, A.; Sukura, A.; Vuorela, P.; Savijoki, K. Alpha-and β-casein components of host milk induce biofilm formation in the mastitis bacterium Streptococcus uberis. Vet. Microbiol. 2011, 149, 381–389.
- Reinoso, E.B. Bovine mastitis caused by streptococcus uberis. Virulence factors and biofilm. J. Microb. Biochem. Technol. 2017.
- Shukla, R.; Pandey, V.; Vadnere, G.P.; Lodhi, S. Role of flavonoids in management of inflammatory disorders. In Bioactive Food as Dietary Interventions for Arthritis and Related Inflammatory Disease; Elsevier: Amsterdam, The Netherlands, 2019; pp. 293–322.
- Alves-Barroco, C.; Roma-Rodrigues, C.; Balasubramanian, N.; Guimarães, M.A.; Ferreira-Carvalho, B.T.; Muthukumaran, J.; Nunes, D.; Fortunato, E.; Martins, R.; Santos-Silva, T.; et al. Biofilm development and computational screening for new putative inhibitors of a homolog of the regulatory protein BrpA in Streptococcus dysgalactiae subsp. dysgalactiae. Int. J. Med. Microbiol. 2019, 309, 169–181.
- Calvinho, L.F.; Almeida, R.A.; Oliver, S.P. Potential virulence factors of Streptococcus dysgalactiae associated with bovine mastitis. Vet. Microbiol. 1998, 61, 93–110.
- Emaneini, M.; Khoramian, B.; Jabalameli, F.; Abani, S.; Dabiri, H.; Beigverdi, R. Comparison of virulence factors and capsular types of Streptococcus agalactiae isolated from human and bovine infections. Microb. Pathog. 2016, 91, 1–4.
- Mudzana, R.; Mavenyengwa, R.T.; Gudza-Mugabe, M. Analysis of virulence factors and antibiotic resistance genes in group B streptococcus from clinical samples. BMC Infect. Dis. 2021, 21, 125.
- Loures, R.A.; de Pádua Pereira, U.; de Carvalho-Castro, G.d.A.; Mian, G.F.; da Costa Custódio, D.A.; da Silva, J.R.; da Costa, G.M. Genetic diversity and virulence genes in Streptococcus uberis strains isolated from bovine mastitis. Semin. Ciências Agrárias 2017, 38, 2595–2605.
- Lämmler, C.; Frede, C. Binding of immunoglobulin G and albumin to Streptococcus dysgalactiae. Zent. Bakteriol. 1989, 271, 321–329.
- Jonsson, H.; Frykberg, L.; Rantamäki, L.; Guss, B. MAG, a novel plasma protein receptor from Streptococcus dysgalactiae. Gene 1994, 143, 85–89.
- Valentin-Weigand, P.; Traore, M.Y.; Blobel, H.; Chhatwal, G.S. Role of α2-macroglobulin in phagocytosis of group A and C streptococci. FEMS Microbiol. Lett. 1990, 70, 321–324.
- Bobadilla, F.J.; Novosak, M.G.; Cortese, I.J.; Delgado, O.D.; Laczeski, M.E. Prevalence, serotypes and virulence genes of Streptococcus agalactiae isolated from pregnant women with 35–37 weeks of gestation. BMC Infect. Dis. 2021, 21, 73.
- Pang, M.; Sun, L.; He, T.; Bao, H.; Zhang, L.; Zhou, Y.; Zhang, H.; Wei, R.; Liu, Y.; Wang, R. Molecular and virulence characterization of highly prevalent Streptococcus agalactiae circulated in bovine dairy herds. Vet. Res. 2017, 48, 1–12.
- International Dairy Federation. Guidelines for the Use and Interpretation of Bovine Milk Somatic Cell Count; International Dairy Federation (IDF/FIL): Schaerbeek, Belgium, 2013; pp. 2–8.
- Blagitz, M.G.; Souza, F.N.; Batista, C.F.; Azevedo, L.F.; Benites, N.R.; Melville, P.A.; Diniz, S.A.; Silva, M.X.; Haddad, J.P.; Heinnemann, M.B.; et al. The neutrophil function and lymphocyte profile of milk from bovine mammary glands infected with Streptococcus dysgalactiae. J. Dairy Res. 2015, 82, 460–469.
- Beecher, C.; Daly, M.; Ross, R.P.; Flynn, J.; McCarthy, T.V.; Giblin, L. Characterization of the bovine innate immune response in milk somatic cells following intramammary infection with Streptococcus dysgalactiae subspecies dysgalactiae. J. Dairy Sci. 2012, 95, 5720–5729.
- Florindo, C.; Barroco, C.A.; Silvestre, I.; Damião, V.; Gomes, J.P.; Spellerberg, B.; Santos-Sanches, I.; Borrego, M.J. Capsular Type, Sequence Type and Microbial Resistance Factors Impact on DNase Activity of Streptococcus agalactiae Strains from Human and Bovine Origin. Eur. J. Microbiol. Immunol. 2018, 8, 149–154.
- Smeesters, P.R.; McMillan, D.J.; Sriprakash, K.S. The streptococcal M protein: A highly versatile molecule. Trends Microbiol. 2010, 18, 275–282.
- Vasi, J.; Frykberg, L.; Carlsson, L.E.; Lindberg, M.; Guss, B. M-like proteins of Streptococcus dysgalactiae. Infect. Immun. 2000, 68, 294–302.
- Duarte, R.S.; Bellei, B.C.; Miranda, O.P.; Brito, M.A.V.P.; Teixeira, L.M. Distribution of antimicrobial resistance and virulence-related genes among Brazilian group B streptococci recovered from bovine and human sources. Antimicrob. Agents Chemother. 2005, 49, 97–103.
- Chen, P.; Qiu, Y.; Liu, G.; Li, X.; Cheng, J.; Liu, K.; Qu, W.; Zhu, C.; Kastelic, J.P.; Han, B.; et al. Characterization of Streptococcus lutetiensis isolated from clinical mastitis of dairy cows. J. Dairy Sci. 2021, 104, 702–714.
More
Information
Subjects:
Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.6K
Revisions:
2 times
(View History)
Update Date:
05 Aug 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




