Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Xiaogang Li | + 2086 word(s) | 2086 | 2021-07-06 08:16:48 | | | |
| 2 | Ron Wang | Meta information modification | 2086 | 2021-08-03 03:15:56 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Li, X. Cell Cycle Regulation and Ciliogenesis. Encyclopedia. Available online: https://encyclopedia.pub/entry/12665 (accessed on 07 February 2026).
Li X. Cell Cycle Regulation and Ciliogenesis. Encyclopedia. Available at: https://encyclopedia.pub/entry/12665. Accessed February 07, 2026.
Li, Xiaogang. "Cell Cycle Regulation and Ciliogenesis" Encyclopedia, https://encyclopedia.pub/entry/12665 (accessed February 07, 2026).
Li, X. (2021, August 02). Cell Cycle Regulation and Ciliogenesis. In Encyclopedia. https://encyclopedia.pub/entry/12665
Li, Xiaogang. "Cell Cycle Regulation and Ciliogenesis." Encyclopedia. Web. 02 August, 2021.
Copy Citation
Primary cilia biogenesis has been closely associated with cell cycle progression. Cilia assemble when cells exit the cell cycle and enter a quiescent stage at the post-mitosis phase, and disassemble before cells re-enter a new cell cycle. Studies have focused on how the cell cycle coordinates with the cilia assembly/disassembly process, and whether and how cilia biogenesis affects the cell cycle. Appropriate regulation of the functions and/or expressions of ciliary and cell-cycle-associated proteins is pivotal to maintaining bodily homeostasis. Epigenetic mechanisms, including DNA methylation and histone/chromatin modifications, are involved in the regulation of cell cycle progression and cilia biogenesis.
cell cycle
primary cilia
epigenetic regulator
ciliogenesis
1. Introduction
The cell cycle is a complicated and finely tuned process that takes place in a cell as it grows and divides, and is composed of interphase (G1, S, and G2 phases), followed by the mitotic phase (mitosis and cytokinesis), and the G0 (quiescence) phase [1][2]. There are at least two decisive cell cycle points identified in the G1 and G2 phases, whereas the G0/G1 phases seem more variable in different cells, resulting in the variable length of the cell cycle [3]. The regulation of the cell cycle and cell proliferation rates varies in different cells.
The cell cycle is regulated by numerous regulatory proteins, which direct a cell through a series of highly ordered events, culminating in the production of two daughter cells. The central regulators of the cell cycle are cyclin-dependent kinases (CDKs), which form complexes with the corresponding cyclins in different phases to promote cell cycle progression (Figure 1) [4][5][6][7]. The activity of CDKs can be inhibited by their canonical inhibitors—including p16, p21, and p27—via direct interaction with CDK–cyclin complexes [8][9]. Recent studies indicate that epigenetic mechanisms are involved in the regulation of cell cycle progression and the expression of CDKs (Figure 1).
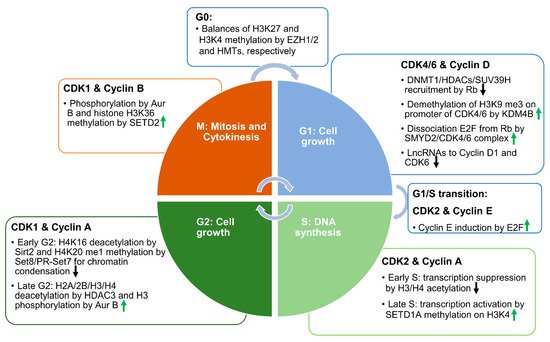
Figure 1. The associations between CDKs, cyclins, and the epigenetic mechanisms in each phase of the cell cycle. Schematic diagram showing the functions of different CDK–cyclin pairs in each phase of the cell cycle. In the G1 phase, CDK4 or CDK6 coupled with cyclin D phosphorylates and removes Rb from the Rb–E2F complex to facilitate E2F-mediated transcription of cell cycle effectors. CDK4 and CDK6 interact with a histone methyltransferase—SMYD2—to regulate its activity. SMYD2 methylation of Rb may facilitate the phosphorylation of Rb by CDK4/6. In addition, SMYD2 directly regulates the transcription of CDK4 and CDK6. In the S phase, CDK2 coupled with cyclin A or E reinforces the phosphorylation of Rb to promote DNA synthesis. In the early S phase, the acetylation of histones suppresses transcription, while later in the S phase, SETD1A-mediated histone methylation reactivates transcription, and promotes the transition from the S phase to the G2 phase. In the G2 phase, CDK1 coupled with cyclin A or B phosphorylates histone tails at different sites to promote mitotic entry. HDAC3-mediated histone deacetylation is involved in this process. In the meantime, SIRT2-mediated deacetylation of histone H4K16 and SET8/PR-SET7-mediated methylation of H4K20 synergistically regulate cell cycle progression. Eventually, when histone H3Ser10 is globally phosphorylated, cells enter the mitosis phase. In the M phase, SETD2-mediated tubulin and histone methylation promotes cytokinesis and the separation of cells into two daughter cells. In the G0 phase, histone methyltransferase (HMT)-mediated H3K4 trimethylation and EZH1/2 -mediated H3K27 trimethylation dynamically control the chromatin status. Upward-pointing green arrows represent the propelling effects on the cell cycle. Downward-pointing black arrows represent the inhibiting effects on the cell cycle.
The cilium is a cell-cycle-associated organelle, in that cilia formation is dynamically coordinated with cell cycle progression. The cilium initiates its assembly at the G0 or G1 phase and elongates through the S phase until centrosomes commence duplication, at which point cilia resorption progresses to completion before the cells begin mitosis. After mitosis, when cells re-enter the cell cycle, centrosomes are again able to assemble primary cilia at either the G0 or early G1 phase [10][11]. Factors that repress the transition from the late M phase to the G1 phase or G1/S entry—such as the inhibitors of CDK2, CDK4, CDK6, and their corresponding cyclins—would induce the initiation of cilia. In contrast, factors that promote cell cycle to restart from the post-M phase would inhibit cilia formation [12][13]. Thus, the canonical understanding is that the cell cycle plays a predominant role in the regulation of cilia biogenesis. However, recent studies indicate that ciliary assembly proteins can also regulate cell cycle progression, thereby shedding light on the effects of ciliogenesis on cell cycle progression. To further understand the relationship between the cell cycle and ciliogenesis, it is necessary to clarify how the cell cycle and ciliogenesis communicate with one another, and coordinate their processes to an appropriate degree in order to maintain cellular hemostasis.
Recent studies suggest that the cell cycle and cilia biogenesis can be regulated by epigenetic mechanisms [12][14]. Epigenetics is the study of heritable genome-wide changes in phenotype or gene expression caused by mechanisms other than changes in the underlying DNA sequence [15]. Epigenetic mechanisms include DNA methylation, histone modifications, and noncoding-RNA-associated silencing [16]. Growing evidence indicates that there is a well-organized network among cell cycle, ciliogenesis, and epigenetic regulators, in which epigenetic regulators are not only involved in the regulation of the cell cycle and ciliogenesis, but can in turn be regulated by cell cycle regulators and contribute to bodily homeostasis. This review will mainly focus on the current understanding of epigenetic mechanisms in the regulation of the cell cycle and ciliogenesis. In particular, we will focus on how epigenetic mechanisms regulate the crosstalk between the cell cycle and ciliogenesis. In addition, we will discuss the roles and mechanisms of cilia-dependent signaling pathways in cell cycle regulation.
2. Epigenetic Regulation on Cell Cycle
The process of cell division and cell proliferation is important for the growth and development of an organism, and organs undergo cellular evolution, renewal, or repair [17][18]. To maintain bodily homeostasis, it is essential to sequentially control the molecular events that occur during the cell cycle. Dysregulation of these processes can lead to various diseases, including cancer and developmental deficiency. The mammalian cell cycle is composed of four phases: G1 (the post-mitotic interphase), S (the DNA synthetic phase), G2 (the post-synthetic phase), and M (the mitotic phase) [19]. The G0 phase is usually difficult to distinguish from G1, for which reason it is usually termed the “G0/G1” phase. The cell cycle is regulated by CDKs and their associated cyclins at different stages (Figure 1). Specifically, CDK4/6 complexes with cyclin D at the early G1 phase; CDK2 complexes with cyclin E during the late G1 phase, or the G1/S phase transition, and complexes with cyclin A at the S phase; CDK1 complexes with cyclin A at the G2 phase and cyclin B at the M phase [20]. CDKs can be regulated by the intrinsic regulators, including p53, p21, p16, and cdc25. Epigenetic mechanisms play essential roles in cell cycle regulation, either through controlling the expression of cell-cycle-related genes—such as CDKs—or through the modification of chromatin condensation and controlling the related signaling proteins via histone marks (Figure 1).
3. Epigenetic Regulation on Ciliogenesis
Cilia are microtubule-based structures that protrude from the cell surface. There are two types of cilia—motile, and non-motile cilia—serving motile and sensory functions, respectively [21][22]. In animals, motile cilia are found on limited tissues, such as respiratory epithelium and choroid plexus epithelial cells [23]. Non-motile cilia, also called primary cilia, can be found on nearly every cell type, and function as sensors for mechanical and chemical environmental cues that regulate cellular differentiation or division [24][25]. In this review, we mainly focus on primary cilia, and the term “cilium/cilia” represents the primary cilium/cilia only, unless otherwise specified. In general, the cilium undergoes cycles of assembly and disassembly that are regulated tightly by a complicated protein network, with proteins trafficking from the base to the cilia tip (anterograde), and from the cilia tip back to the cytoplasm (retrograde), mediated by intraflagellar transport (IFT) complexes of the IFT-B and IFT-A families, driven by a kinesin-2 motor in the anterograde direction and a dynein motor in the retrograde direction, respectively [26].
4. Crosstalk between the Cell Cycle and Ciliogenesis
With the knowledge of the process of ciliogenesis, from initiation and elongation to resorption, it is necessary to understand the crosstalk between the cell cycle and ciliogenesis. In general, the cilia assembly and disassembly cycle is closely associated with the cell cycle, as described above, which suggests that the cilia biogenesis process can be passively regulated by cell cycle progression. However, several studies have provided some new clues about the relationship between the primary cilia and the cell cycle. Firstly, studies have indicated that by the inhibition of several negative regulators of ciliogenesis, cilia formation was maintained in proliferating cells and the cell cycle was arrested, while overexpression of these factors inhibited cilia formation, even in serum-starved cells [27]. It has been reported that trichoplein complexed with Aurora A (Aur A) blocks aberrant primary cilia assembly in proliferating cells. Overexpression of trichoplein inhibited primary cilia assembly in serum-starved cells, whereas knockdown of trichoplein induced primary cilia assembly, even in serum-contained medium, and induced G0/G1 arrest [28], suggesting the independence of ciliogenesis from the cell cycle. The following findings also indicate that ciliogenesis can function as a regulator of the cell cycle and cell proliferation: (1) defects of the cilia or lack of cilia cause cellular proliferation abnormalities and diseases; (2) primary cilia sense urine flow, and are essential for the maintenance of the epithelial architecture in the kidneys; and (3) defects of primary cilia cause cystic kidney disease characterized by epithelial abnormalities, suggesting the roles of primary cilia in the regulation of the cell cycle and cell proliferation.
5. Cilia-Dependent Signaling Pathways in Cell Cycle Regulation, and the Association of Epigenetic Mechanisms with These Signaling Pathways
Cilia also function as regulatory switches to control diverse signaling pathways, including Hedgehog-, Wnt-, PDGFR-, Notch-, TGF-β-, mTOR-, and GPCR-associated signaling. All of these signaling pathways play crucial roles in various cellular processes, such as in organ and embryonic development, cardiac function, planar cell polarity, transactivation, differentiation, cell cycle progression, apoptosis, tissue homeostasis, and the immune response. Growing evidence demonstrates that these signaling pathways can be regulated by epigenetic mechanisms in human diseases.
6. Therapeutic Targets of Cell-Cycle- and Ciliogenesis-Associated Epigenetic Regulators
Increasing evidence exhibits the roles of epigenetic regulators in regulating the cell cycle and ciliogenesis during organ development, and in many proliferative diseases (such as cancers) and cilia-deficient diseases (termed as ciliopathies) [29][30][31]. Thus, targeting abnormal epigenetic regulators might be an advanced therapeutic strategy.
The role of the histone methyltransferase EZH2 in driving transformation from benign to malignant melanoma has been associated with the regulation of both the cell cycle and ciliogenesis. On one hand, EZH2 suppresses the expression of ciliary proteins through its repressive histone mark H3K27me3, by binding to the promoters of many ciliary genes, including WDR19, IFT81, FUZ, etc. On the other hand, loss of cilia results in the inappropriate activation of cilia-dependent signaling. In A375 human melanoma cells, the enhanced WNT/β-catenin activity and nuclear accumulation is accompanied by loss of cilia, due to the absence of cilia-mediated spatial segregation. The involvement of EZH2 in this process was evidenced by the findings that increased EZH enhances WNT/β-catenin signaling in benign melanocytic cells, and inhibition of EZH2 with GSK503 induces cilia assembly in most melanoma cell lines. Blockade of EZH induces cilia assembly, which could be reversed by silencing ciliary genes [31]. The EZH2 inhibitor GSK503 is a small molecule, which exerts its inhibition by competition with S-adenosylmethionine (SAM), the donor of the methyl group. There are several other EZH2 inhibitors—such as DZNep, EPZ005687, EI1, GSK126, UNC1999, GSK503, and EPZ-6438—which are all small molecules and function as SAM-competitive inhibitors. DZNep is one of the purine nucleoside analogs, which has been tested in the treatment of hematological malignancies and autoimmune diseases. EPZ005687 is a selective inhibitor of EZH2 rather than EZH1. EI1 mainly regulates and activates EZH2 target genes via the removal of repressive histone mark H3K27 methylation. GSK126 has been tested in a live failure murine model, in which it decreased inflammation response through the reduction of H3K27 methylation. UNC1999 functions similarly to GSK126. GSK503 has been tested in a human melanoma murine model. EPZ-6438 is an improved product of EPZ005687. Among those inhibitors, tazemetostat (EPZ-6438) has been approved for the treatment of epithelioid sarcoma [32]. Whether those EZH2 inhibitors also affect ciliogenesis in different disease models needs to be further investigated.
References
- Schafer, K.A. The cell cycle: A review. Vet. Pathol. 1998, 35, 461–478.
- Chao, H.X.; Fakhreddin, R.I.; Shimerov, H.K.; Kedziora, K.M.; Kumar, R.J.; Perez, J.; Limas, J.C.; Grant, G.D.; Cook, J.G.; Gupta, G.P.; et al. Evidence that the human cell cycle is a series of uncoupled, memoryless phases. Mol. Syst. Biol. 2019, 15, e8604.
- Matson, J.P.; Cook, J.G. Cell cycle proliferation decisions: The impact of single cell analyses. FEBS J. 2017, 284, 362–375.
- Wang, J.; Yang, T.; Xu, G.; Liu, H.; Ren, C.; Xie, W.; Wang, M. Cyclin-Dependent Kinase 2 Promotes Tumor Proliferation and Induces Radio Resistance in Glioblastoma. Transl. Oncol. 2016, 9, 548–556.
- Sviderskiy, V.O.; Blumenberg, L.; Gorodetsky, E.; Karakousi, T.R.; Hirsh, N.; Alvarez, S.W.; Terzi, E.M.; Kaparos, E.; Whiten, G.C.; Ssebyala, S.; et al. Hyperactive CDK2 Activity in Basal-like Breast Cancer Imposes a Genome Integrity Liability that Can Be Exploited by Targeting DNA Polymerase epsilon. Mol. Cell 2020, 80, 682–698 e687.
- Onesti, C.E.; Jerusalem, G. CDK4/6 inhibitors in breast cancer: Differences in toxicity profiles and impact on agent choice. A systematic review and meta-analysis. Expert Rev. Anticancer 2021, 21, 283–298.
- Servetto, A.; Napolitano, F.; De Angelis, C.; De Placido, P.; Giuliano, M.; Arpino, G.; De Placido, S.; Bianco, R.; Formisano, L. A review of the use of next generation sequencing methodologies to identify biomarkers of resistance to CDK4/6 inhibitors in ER+/HER2- breast cancer. Crit. Rev. Oncol. Hematol. 2021, 157, 103191.
- Duronio, R.J.; Xiong, Y. Signaling pathways that control cell proliferation. Cold Spring Harb. Perspect. Biol. 2013, 5, a008904.
- Besson, A.; Dowdy, S.F.; Roberts, J.M. CDK inhibitors: Cell cycle regulators and beyond. Dev. Cell 2008, 14, 159–169.
- Pan, J.; Snell, W. The primary cilium: Keeper of the key to cell division. Cell 2007, 129, 1255–1257.
- Plotnikova, O.V.; Pugacheva, E.N.; Golemis, E.A. Primary cilia and the cell cycle. Methods Cell Biol. 2009, 94, 137–160.
- Li, L.X.; Zhou, J.X.; Wang, X.; Zhang, H.; Harris, P.C.; Calvet, J.P.; Li, X. Cross-talk between CDK4/6 and SMYD2 regulates gene transcription, tubulin methylation, and ciliogenesis. Sci. Adv. 2020, 6.
- Hu, S.; Danilov, A.V.; Godek, K.; Orr, B.; Tafe, L.J.; Rodriguez-Canales, J.; Behrens, C.; Mino, B.; Moran, C.A.; Memoli, V.A.; et al. CDK2 Inhibition Causes Anaphase Catastrophe in Lung Cancer through the Centrosomal Protein CP110. Cancer Res. 2015, 75, 2029–2038.
- Pugacheva, E.N.; Jablonski, S.A.; Hartman, T.R.; Henske, E.P.; Golemis, E.A. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 2007, 129, 1351–1363.
- Devaskar, S.U.; Raychaudhuri, S. Epigenetics--a science of heritable biological adaptation. Pediatr. Res. 2007, 61, 1R–4R.
- Li, X. Epigenetics and cell cycle regulation in cystogenesis. Cell. Signal. 2020, 68, 109509.
- McGill, C.J.; Brooks, G. Cell cycle control mechanisms and their role in cardiac growth. Cardiovasc. Res. 1995, 30, 557–569.
- Stein, G.S.; Stein, J.L.; van Wijnen, A.; Lian, J.B.; Quesenberry, P.J. Molecular mechanisms mediating control of cell cycle and cell growth. Exp. Hematol. 1995, 23, 1053–1061.
- Vermeulen, K.; Van Bockstaele, D.R.; Berneman, Z.N. The cell cycle: A review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003, 36, 131–149.
- Malumbres, M.; Barbacid, M. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer 2009, 9, 153–166.
- Pazour, G.J.; Witman, G.B. The vertebrate primary cilium is a sensory organelle. Curr. Opin. Cell Biol. 2003, 15, 105–110.
- Jain, R.; Javidan-Nejad, C.; Alexander-Brett, J.; Horani, A.; Cabellon, M.C.; Walter, M.J.; Brody, S.L. Sensory functions of motile cilia and implication for bronchiectasis. Front. Biosci. (Sch. Ed.) 2012, 4, 1088–1098.
- Ringers, C.; Olstad, E.W.; Jurisch-Yaksi, N. The role of motile cilia in the development and physiology of the nervous system. Philos Trans. R Soc. Lond B Biol. Sci. 2020, 375, 20190156.
- Singh, M.; Chaudhry, P.; Merchant, A.A. Primary cilia are present on human blood and bone marrow cells and mediate Hedgehog signaling. Exp. Hematol. 2016, 44, 1181–1187 e1182.
- Hua, K.; Ferland, R.J. Primary cilia proteins: Ciliary and extraciliary sites and functions. Cell Mol. Life Sci. 2018, 75, 1521–1540.
- Rosenbaum, J.L.; Witman, G.B. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 2002, 3, 813–825.
- Goto, H.; Inaba, H.; Inagaki, M. Mechanisms of ciliogenesis suppression in dividing cells. Cell Mol. Life Sci. 2017, 74, 881–890.
- Inoko, A.; Matsuyama, M.; Goto, H.; Ohmuro-Matsuyama, Y.; Hayashi, Y.; Enomoto, M.; Ibi, M.; Urano, T.; Yonemura, S.; Kiyono, T.; et al. Trichoplein and Aurora A block aberrant primary cilia assembly in proliferating cells. J. Cell Biol. 2012, 197, 391–405.
- Li, L.X.; Fan, L.X.; Zhou, J.X.; Grantham, J.J.; Calvet, J.P.; Sage, J.; Li, X. Lysine methyltransferase SMYD2 promotes cyst growth in autosomal dominant polycystic kidney disease. J. Clin. Investig. 2017, 127, 2751–2764.
- Zhou, X.; Fan, L.X.; Li, K.; Ramchandran, R.; Calvet, J.P.; Li, X. SIRT2 regulates ciliogenesis and contributes to abnormal centrosome amplification caused by loss of polycystin-1. Hum. Mol. Genet. 2014, 23, 1644–1655.
- Zingg, D.; Debbache, J.; Pena-Hernandez, R.; Antunes, A.T.; Schaefer, S.M.; Cheng, P.F.; Zimmerli, D.; Haeusel, J.; Calcada, R.R.; Tuncer, E.; et al. EZH2-Mediated Primary Cilium Deconstruction Drives Metastatic Melanoma Formation. Cancer Cell 2018, 34, 69–84 e14.
- Lim, H.J.; Kim, M. EZH2 as a Potential Target for NAFLD Therapy. Int. J. Mol. Sci. 2020, 21, 8617.
More
Information
Subjects:
Cell Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
957
Revisions:
2 times
(View History)
Update Date:
03 Aug 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




