
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Judit Ovádi | + 1182 word(s) | 1182 | 2021-07-29 07:48:07 | | | |
| 2 | Amina Yu | Meta information modification | 1182 | 2021-08-02 07:41:16 | | |
Video Upload Options
The cytoskeleton is an intricate network of filaments running through the cytoplasm that helps cells to maintain their shape and internal organization. Coronaviruses such as SARS-CoV-2 are intimately involved in cytoskeleton-related physiological and pathological processes. The discovery of the exact mechanisms of the subversion of host cells by SARS-CoV-2 is critical to the validation of specific drug targets and effective treatments.
1. Overview: Coronaviruses and the Cytoskeleton
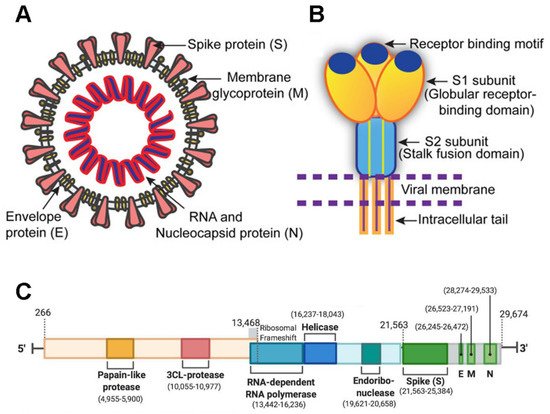
SARS-CoV-2 is a member of the Coronaviridae family and has a large 29,903-nucleotide, positive-strand RNA genome [1]. The genome organization of SARS-CoV-2 is similar to other CoVs such as HCoV-OC43, MERS-CoV, SARS-CoV, HCoV-229E, and HCoV-NL63. One third of its genome encodes accessory and structural proteins, which includes the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins [2][3] (green blocks in Figure 1 ); the other two thirds of the genome are occupied by two large overlapping open reading frames (ORF1a and ORF1b) that are translated into polyproteins and are processed to generate 16 non-structural proteins. A ribosomal frameshift is located between ORF1a and ORF1b. The non-structural proteins include papain-like protease, 3CL-protease, RNA-dependent RNA polymerase, helicase, endoribonuclease, and viroporins.
The cytoskeleton, which plays a key role in several viral processes, is an intricate network of filaments running through the cytoplasm that helps cells to maintain their shape and internal organization ( Figure 2 ). This is achieved via the dynamics of the actin filaments (AFs), microtubule filaments (MFs), and intermediate filaments (IFs) that constitute the cytoskeletal network [4][5]; in particular, the cytoskeleton provides the mechanical support that enables cells to perform many vital functions.
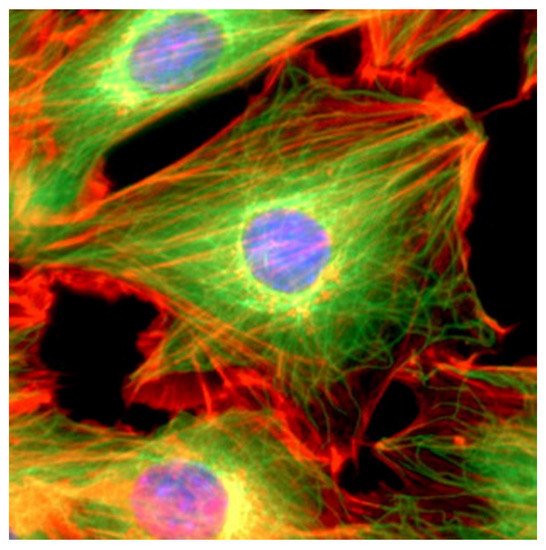
These functions include cargo transport, signal transduction, and the control of cell shape, movement, and division. Recent studies have shown the importance of the interactions between coronaviruses and cytoskeletal filaments. The entry of the virus involves IFs and MFs, which is followed by the use of MFs for transportation to replication and assembly sites, and the harnessing of the polymerization of AFs to force release [1].
2. Cytoskeletal Transport of Influenza Viruses
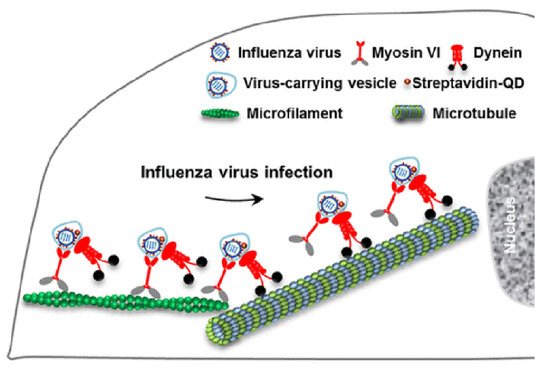
Viral homologs of host proteins can mimic fundamental cell processes during the course of the viral life cycle [7]. The well-characterized influenza virus is often used as a model system for understanding viral infections [8]. When infecting host cells, the virus must move along AFs at the cell periphery and then move along MFs through the cytosol to reach the perinuclear region for genome release [9]. Within the host cells, myosin VI (MyoVI) and dynein are responsible for virus transport on the AFs and MFs with the two motor proteins being attached to the same virus-carrying vesicle ( Figure 3 ). MyoVI drives viruses along AFs with dynein as a passenger on the vesicle, and then dynein drives the viruses along MFs with MyoVI as a passenger. It has been revealed that the “driver switchover” mechanism from AFs to MFs enables the successful transport of the virus within the host cell [9].
3. Microtubule Filaments and Coronaviruses
In many cell types, cytoplasmic dynein motors transport cargoes in a retrograde manner toward the minus end of MTs, which are frequently anchored at the MicroTubule Organizing Center (MTOC) [10]. The long-range transport of the coronaviruses to and from the cell periphery is mediated by dynein and kinesin on the MFs, while the actin and myosin filaments mediate the short-range transport [11].
CoV infections of cells may stimulate the formation of autophagosomes, which have double membranes (see Section 6 ); this formation is facilitated by MFs on which their subsequent movement and fusion depend [12]. In fact, CoV infections are considered to entail processes closely associated with autophagy, which may even promote CoV infection and replication ([13] and references therein). However, the interplay between the autophagy machinery and the CoVs including the SARS-CoV-2 is unclear so far [14].
The trafficking of viruses on the MT network by dynein and kinesin-1 motors plays an important role in the replication and spread of many viruses, as supported by excellent studies carried out with Porcine Epidemic Diarrhea Virus (PEDV), a member of the Coronavirus family [15]. Using a single-virus tracking technique, the molecular mechanisms of the transport of PEDV have revealed the involvement of this trafficking machinery in the fusion of the virus with the membrane and in its accumulation in the perinuclear region. The dynamically monitored intracellular transport displayed different mechanisms such as unidirectional movements toward MT plus/minus ends as well as bidirectional movements along different MTs. The findings of these studies have greatly contributed to our understanding of the pathogenesis of CoVs.
Finally, in the context of MFs, cilia should also be considered. Cilia are composite structures based on MTs and present on the cell surface [16]. Ciliopathies are associated with a wide range of clinical features that include chronic respiratory problems. It is therefore significant that structural damage to the respiratory epithelium and abnormal ciliary function are typical pathological symptoms of CoV infection with different CoVs causing cilia loss (via changes to the structure of MFs) and/or severe damage in the upper respiratory tract and lung [16].
4. Actin Filaments and Coronaviruses
After binding to the host cell, viruses can use the depolymerization of AFs to “surf” to their entry sites where further dynamic rearrangements of AFs are involved in virus internalization [1]. On one hand, cofilin, a host protein that promotes AF depolymerization, is phosphorylated by some coronaviruses to inhibit this depolymerization and assist with entry [17]. On the other hand, ezrin, a host protein that links the membrane to the actin cytoskeleton, can bind to the C-terminus of the Spike protein and inhibit the entry and fusion of SARS-CoV [18]. In the case of antibody binding to Feline Infectious Peritonitis Virus-infected monocytes, internalization is initiated and driven by Myo I and Myosin Light Chain Kinase, whilst subsequent passage through the cortical actin barrier involves Myo VI [11]. At the later stage of the synthesis of the viral proteins and genomes, AFs retract from the plasma membrane to form a ring associated with the nucleus and to bind to virus particles near the nuclear membrane [1]. The disruption of AF dynamics counteracts the actin ring formation and virus replication. It should be noted that actin rearrangements can result from the action of the N protein of SARS-CoV, which induces the p38 mitogen-activated protein kinase cascade [19]. The viral structural proteins move to the ER-Golgi intermediate compartment where, at least in one coronavirus, an AF-crosslinking protein, filamin A, interacts with the Spike protein [20]. Finally, interaction between β-actin and the M protein is essential for the assembly and budding of Infectious Bronchitis Virus [21], whilst the thickening of AFs below the cell surface is proposed to provide the bending force to extrude SARS-CoV particles [22].
References
- Wen, Z.; Zhang, Y.; Lin, Z.; Shi, K.; Jiu, Y. Cytoskeleton-a crucial key in host cell for coronavirus infection. J. Mol. Cell Biol. 2021, 12, 968–979.
- Mittal, A.; Manjunath, K.; Ranjan, R.K.; Kaushik, S.; Kumar, S.; Verma, V. COVID-19 pandemic: Insights into structure, function, and hACE2 receptor recognition by SARS-CoV-2. PLoS Pathog. 2020, 16, e1008762.
- Chilamakuri, R.; Agarwal, S. COVID-19: Characteristics and Therapeutics. Cells 2021, 10, 206.
- Conde, C.; Caceres, A. Microtubule assembly, organization and dynamics in axons and dendrites. Nat. Rev. Neurosci. 2009, 10, 319–332.
- de Forges, H.; Bouissou, A.; Perez, F. Interplay between microtubule dynamics and intracellular organization. Int. J. Biochem. Cell Biol. 2012, 44, 266–274.
- Suresh, S. Biomechanics and biophysics of cancer cells. Acta Biomater. 2007, 3, 413–438.
- Kalicharran, K.; Dales, S. Involvement of microtubules and the microtubule-associated protein tau in trafficking of JHM virus and components within neurons. Adv. Exp. Med. Biol. 1995, 380, 57–61.
- Otter, J.A.; Donskey, C.; Yezli, S.; Douthwaite, S.; Goldenberg, S.D.; Weber, D.J. Transmission of SARS and MERS coronaviruses and influenza virus in healthcare settings: The possible role of dry surface contamination. J. Hosp. Infect. 2016, 92, 235–250.
- Zhang, L.J.; Xia, L.; Liu, S.L.; Sun, E.Z.; Wu, Q.M.; Wen, L.; Zhang, Z.L.; Pang, D.W. A “Driver Switchover” Mechanism of Influenza Virus Transport from Microfilaments to Microtubules. Acs Nano. 2018, 12, 474–484.
- Hook, P.; Vallee, R.B. The dynein family at a glance. J. Cell Sci. 2006, 119, 4369–4371.
- Dewerchin, H.L.; Desmarets, L.M.; Noppe, Y.; Nauwynck, H.J. Myosins 1 and 6, myosin light chain kinase, actin and microtubules cooperate during antibody-mediated internalisation and trafficking of membrane-expressed viral antigens in feline infectious peritonitis virus infected monocytes. Vet. Res. 2014, 45, 17.
- Kochl, R.; Hu, X.W.; Chan, E.Y.; Tooze, S.A. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic 2006, 7, 129–145.
- Zhao, Z.; Lu, K.; Mao, B.; Liu, S.; Trilling, M.; Huang, A.; Lu, M.; Lin, Y. The interplay between emerging human coronavirus infections and autophagy. Emerg. Microbes Infect. 2021, 10, 196–205.
- Bernasconi, R.; Noack, J.; Molinari, M. Unconventional roles of nonlipidated LC3 in ERAD tuning and coronavirus infection. Autophagy 2012, 8, 1534–1536.
- Hou, W.; Kang, W.; Li, Y.; Shan, Y.; Wang, S.; Liu, F. Dynamic Dissection of Dynein and Kinesin-1 Cooperatively Mediated Intercellular Transport of Porcine Epidemic Diarrhea Coronavirus along Microtubule Using Single Virus Tracking. Virulence 2021, 12, 615–629.
- Buqaileh, R.; Saternos, H.; Ley, S.; Aranda, A.; Forero, K.; AbouAlaiwi, W.A. Can cilia provide an entry gateway for SARS-CoV-2 to human ciliated cells? Physiol. Genom. 2021, 53, 249–258.
- Lv, X.; Li, Z.; Guan, J.; Hu, S.; Zhang, J.; Lan, Y.; Zhao, K.; Lu, H.; Song, D.; He, H.; et al. Porcine Hemagglutinating Encephalomyelitis Virus Activation of the Integrin alpha5beta1-FAK-Cofilin Pathway Causes Cytoskeletal Rearrangement to Promote Its Invasion of N2a Cells. J. Virol. 2019, 93.
- Millet, J.K.; Kien, F.; Cheung, C.Y.; Siu, Y.L.; Chan, W.L.; Li, H.; Leung, H.L.; Jaume, M.; Bruzzone, R.; Peiris, J.S.; et al. Ezrin interacts with the SARS coronavirus Spike protein and restrains infection at the entry stage. PLoS ONE 2012, 7, e49566.
- Surjit, M.; Liu, B.; Jameel, S.; Chow, V.T.; Lal, S.K. The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochem. J. 2004, 383, 13–18.
- Trincone, A.; Schwegmann-Wessels, C. Looking for a needle in a haystack: Cellular proteins that may interact with the tyrosine-based sorting signal of the TGEV S protein. Virus Res. 2015, 202, 3–11.
- Wang, J.; Fang, S.; Xiao, H.; Chen, B.; Tam, J.P.; Liu, D.X. Interaction of the coronavirus infectious bronchitis virus membrane protein with beta-actin and its implication in virion assembly and budding. PLoS ONE 2009, 4, e4908.
- Ng, M.L.; Lee, J.W.; Leong, M.L.; Ling, A.E.; Tan, H.C.; Ooi, E.E. Topographic changes in SARS coronavirus-infected cells at late stages of infection. Emerg. Infect. Dis. 2004, 10, 1907–1914.




