Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Yoshiaki Kamiyama | + 2744 word(s) | 2744 | 2021-07-24 17:16:01 | | | |
| 2 | Conner Chen | Meta information modification | 2744 | 2021-07-26 07:46:48 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Kamiyama, Y. SnRK2 in Plants. Encyclopedia. Available online: https://encyclopedia.pub/entry/12407 (accessed on 07 February 2026).
Kamiyama Y. SnRK2 in Plants. Encyclopedia. Available at: https://encyclopedia.pub/entry/12407. Accessed February 07, 2026.
Kamiyama, Yoshiaki. "SnRK2 in Plants" Encyclopedia, https://encyclopedia.pub/entry/12407 (accessed February 07, 2026).
Kamiyama, Y. (2021, July 26). SnRK2 in Plants. In Encyclopedia. https://encyclopedia.pub/entry/12407
Kamiyama, Yoshiaki. "SnRK2 in Plants." Encyclopedia. Web. 26 July, 2021.
Copy Citation
Reversible phosphorylation is a major mechanism for regulating protein function and controls a wide range of cellular functions including responses to external stimuli. The plant-specific SNF1-related protein kinase 2s (SnRK2s) function as central regulators of plant growth and development, as well as tolerance to multiple abiotic stresses.
abscisic acid
osmotic stress
protein kinase
growth promotion
SnRK2
1. Introduction
Throughout their lifecycles, plants are sometimes exposed to water-deficit conditions such as drought, salinity and freezing. Such conditions are perceived by plant tissues as an osmotic stress that reduces plant growth, leading to losses in crop yield worldwide. To cope with such conditions and maintain cellular homeostasis, plant cells have developed physiological and molecular mechanisms to recognize and respond to external stresses in a precise and timely manner. Such external stimuli are generally perceived at the plasma membrane of cells and then transmitted to the entire cell through various signaling pathways. Protein phosphorylation is a post-translational modification that plays an important role in the cellular response to osmotic stress. Phosphorylation is catalyzed by protein kinases, which transfer a γ- phosphoryl group from ATP to specific serine, threonine, tyrosine or sometimes histidine residues within their substrate proteins [1]. Phosphorylation can affect the stability, catalytic activity and subcellular localization of proteins, as well as interactions with other regulatory components [2][3]. This modification is considered reversible because phosphorylated proteins can also be readily dephosphorylated by protein phosphatases [4]. Of the ~28,000 protein-encoding genes in the model plant Arabidopsis thaliana [5], over 1000 genes encode protein kinases [6] and only a small subset of these genes have been functionally characterized to date.
The collective effort by many researchers over the past two decades has resulted in a deeper understanding of the essential roles of a phytohormone abscisic acid (ABA) and SnRK2-type protein kinases in the plant responses to diverse environmental stimuli, especially to osmotic stress [7][8]. ABA plays crucial roles in plant responses to osmotic stress and desiccation, such as stomatal closure, gene expression, seed maturation and dormancy [9][10]. When exposed to water-deficit conditions, plants accumulate higher levels of ABA, which in turn induces protective stress responses via ABA-dependent or -independent pathways, both of which trigger activation of multiple SnRK2s [11][12][13]. Following their activation, SnRK2s transduce the stress signal through phosphorylation of various downstream substrates, including transcription factors and ion channels. In turn, these downstream substrates facilitate protective stress responses [14][15]. Although the mechanisms by which ABA induces SnRK2 activation are well characterized [16][17][18][19], how osmotic stress induces direct activation of SnRK2s was, until recently, less clear. A breakthrough came in 2015, when a pioneering study identified a group B3 Raf-like protein kinase as a direct upstream activator of SnRK2s in the moss [20]. Following this initial study, several additional studies revealed the essential roles of group B Raf-like protein kinases in ABA-independent SnRK2 activation in response to osmotic stress responses in Arabidopsis [21][22][23][24]. In addition to these new connections between SnRK2s and Raf-like kinases, recent data shows that SnRK2-mediated signaling also regulates target of rapamycin (TOR), a master regulator that orchestrates cell proliferation and growth by integrating nutrient, energy, hormone and diverse stress stimuli [25][26][27]. Together, these studies indicate that SnRKs play a pivotal role in the balance between plant growth and stress responses.
2. Role of SnRK2 Protein Kinases in Plant Abiotic Stress Tolerance
SnRK2-type protein kinases belong to the SnRK family and are classified into three subgroups (SnRK1, SnRK2 and SnRK3) according to their sequence similarity and C-terminal domain structure [28] (Figure 1). The heterotrimeric SnRK1s are the most closely related to yeast SUCROSE NON-FERMENTING 1 (SNF1) kinase and mammalian AMP-ACTIVATED PROTEIN KINASEs (AMPKs) [28] and are involved in cellular responses to starvation or nutrient signals [29]. Differing from SnRK1, the SnRK2 and SnRK3 groups are unique to plants. SnRK3 is also named as CALCINEURIN B-LIKE (CBL)-INTERACTING PROTEIN KINASE (CIPK) according to their ability of interacting with Ca2+ sensor CBL proteins, or PROTEIN KINASE S (PKS). The most characterized SnRK3 kinase is SALT OVERLY SENSITIVE 2 (SOS2), also known as SnRK3.11/CIPK24, that is a serine/threonine kinase and that forms a complex with CBL4/SOS3. The SOS2-SOS3 complex acts in the SOS pathway that is required for salt tolerance [30]. SnRK2 families are evolutionarily conserved in major plant lineages, including algae (e.g., Klebsormidium nitens and Chlamydomonas reinhardtii), liverwort (Marchantia polymorpha), moss (Physcomitrella patens), lycophytes (Selaginella moellendorffii) and land plants (Arabidopsis thaliana and Oryza sativa) [31][32][33]. The first reported SnRK2 was PROTEIN KINASE ABA 1 (PKABA1) from wheat and its transcript was induced by ABA [34]. Later, ABA-ACTIVATED PROTEIN KINASE (AAPK) was found from fava bean (Vicia fava) [35] and its Arabidopsis ortholog OPEN STOMATA 1 (OST1)/ SRK2E/ SnRK2.6 was identified [36][37].
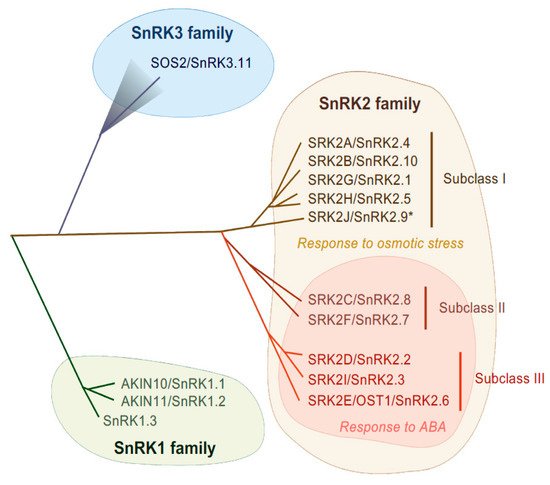
Figure 1. Phylogenetic tree of SnRK family in Arabidopsis. In Arabidopsis, SnRK1, SnRK2 and SnRK3 contain 3, 10 and 25 genes, respectively. SnRK2 family is classified into three subclasses, subclass I, II and III. SnRK2 kinases are activated in response to osmotic stress, except for SRK2J (indicated by *). Subclass II and Subclass III SnRK2s are activated in response to ABA.
SnRK2s are monomeric serine/threonine protein kinases with a molecular mass of about 40 kDa [38], consisting of an N-terminal protein kinase domain and a C-terminal domain that contains stretches of acidic amino acids (acidic patch). The C-terminal domains are further functionally categorized as domain I and domain II, also known as SnRK2-box or ABA-box, respectively [11][39][40]. The amino acid sequences encoding the N-terminal kinase domain are highly conserved among all SnRK2 members, while sequences encoding the C-terminal domain are more divergent [11][28][36]. These C-terminal domains are required for SnRK2 activation in a stress-specific manner, with the SnRK2-box necessary for osmostress-induced or ABA-independent activation and the ABA-box necessary for ABA-dependent activation [11][13][39]. On the basis of the amino acid sequence similarity and responsiveness to ABA or osmotic stress, the SnRK2 family can be classified into three subclasses: I, II and III [12][13] (Figure 1). Although SnRK2-box is relatively similar in all SnRK2 members, the ABA-box is apparently different among SnRK2 subclasses, especially within the acidic patch region. Indeed, the acidic patch is rich in glutamic acid (E) in subclass I SnRK2s, but rich in aspartic acid (D) in subclass II and III [11]. Although only D-rich SnRK2s (subclass II and III) are activated by ABA, all SnRK2 members except for Arabidopsis SRK2J/SnRK2.9 (subclass I) are activated in response to osmotic stress [11][12][13] (Figure 1). Thus, subclass II and III SnRK2s are classified as ABA-responsive SnRK2s, while subclass I SnRK2s are classified as ABA-unresponsive SnRK2s.
2.1. Role of Arabidopsis Subclass III SnRK2s in Regulating Abiotic Stress Responses
The Arabidopsis genome contains ten members of the SnRK2 family, designated SRK2A to SRK2J [36] or SnRK2.1 to SnRK2.10 [28] (Figure 1). Among these subclasses, subclass III members (SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3) are strongly activated by ABA and osmotic stress treatment and play essential roles in the induction of multiple osmotic stress responses including activation of ABA signaling [12]. SRK2E is expressed in guard cells dominantly and plays a key role in stomatal movement [36][37], while SRK2D and SRK2I are mainly involved in ABA signaling during seed germination, seed dormancy and seedling growth [41]. The triple knockout mutant for subclass III SnRK2s (srk2dei) showed extreme insensitivity to ABA [18][42][43][44], indicating that those three kinases are functionally redundant.
Progress has been made in identifying proteins phosphorylated by subclass III SnRK2s. For example, bZIP-type transcription factors AREB/ABFs (ABA-RESPONSIVE ELEMENT-BINDING PROTEINS/ABRE-BINDING FACTORS) are phosphorylated by SnRK2s to induce ABA- or stress-responsive gene expression [45][46] (Figure 2). In guard cells, SRK2E phosphorylates various membrane-localized proteins to induce stomatal closure and reduce transpiration. Activation of anion channels SLOW ANION CHANNEL-ASSOCIATED 1 (SLAC1) [47][48] and QUICK-ACTIVATING ANION CHANNEL 1 (QUAC1) [49] by SRK2E induces anion efflux through the plasma membrane, leading to activation of outward-rectifying K+ channel GATED OUTWARDLY-RECTIFYING K+ CHANNEL (GORK) [50] and K+ release. Another SRK2E target is the inward-rectifying K+ channel POTASSIUM CHANNEL IN ARABIDOPSIS THALIANA 1 (KAT1). SRK2E phosphorylates and inactivates KAT1 to prevent K+ influx [51]. Additionally, SRK2E phosphorylates a NADPH oxidase RESPIRATORY BURST OXIDASE HOMOLOG F (RbohF) to promote apoplast reactive oxygen species (ROS) production [52]. The accumulation of apoplastic ROS is crucial for the induction of stomatal closure. More recent comparative phosphoproteomic analyses identified additional putative substrates downstream of subclass III SnRK2s [14][15]. Among these substrates, SNRK2-SUBSTRATE 1 (SNS1) negatively regulates ABA signaling at cotyledon greening stage [14], whereas SERRATE (SE) and HYPONASTIC LEAVES 1 (HYL1), core components of miRNA processing complex, are phosphorylated by SnRK2s to modulate miRNA accumulation [15][53]. The global view provided by these phosphoproteomic approaches also revealed [-(R/K)-x-x-(pS/pT)-] and [-(pS/pT)-x-x-x-x-(D/E)-] as the preferred motifs that SnRK2s phosphorylate within their substrates in Arabidopsis [14].
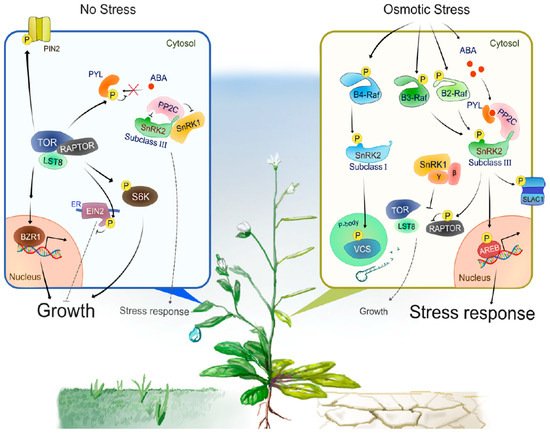
Figure 2. A model of the intracellular signal transduction under optimal growth conditions or during osmotic stress responses.Under optimal growth conditions (No Stress), TOR, RAPTOR and LST8 form a complex and promote cell division, expansion or elongation for plant growth through positive regulation of growth promotion-related factors such as S6K, BZR1 and PIN2. In addition, TOR directly phosphorylates PYLs or EIN2 to negatively regulates stress-associated phytohormone signaling such as ABA and ethylene, respectively. TOR-mediated phosphorylation of PYLs inhibits PYL-binding to ABA, therefore active PP2Cs dephosphorylate and inactivate subclass III SnRK2s to compromise ABA signaling and other protective stress responses. Such a PP2C-SnRK2 complex sequesters SnRK1, thereby allowing TOR activity and growth. Under water-deficit conditions (Osmotic Stress), multiple group B Raf-like protein kinases are rapidly activated. B4 Raf-like protein kinases directly phosphorylate and activate ABA-unresponsive (subclass I) SnRK2s, thereby enhancing mRNA decapping activator VARICOSE (VCS) activity to modulate mRNA population under osmotic stress responses. On the other hand, ABA-responsive (subclass III) SnRK2s activate in the both ABA-dependent and -independent manner. That is, osmotic stress promotes ABA biosynthesis and increased ABA is perceived by PYLs, leading to the formation of PYL-ABA-PP2C complexes, thereby releasing subclass III SnRK2s from inhibition by PP2Cs. In addition, osmostress-activated B2/B3 Raf-like protein kinases directly phosphorylate and activate subclass III SnRK2s. Activated subclass III SnRK2s phosphorylate downstream substrates such as bZIP-type transcription factor AREBs and ion channel SLAC1, to promote ABA responses or other protective stress responses including regulation of stress-responsive gene expression and stomatal closure. SnRK1 is released from the sequestration by PP2C-SnRK2 complex and inhibits TOR-mediated signaling to repress growth. The β and γ represent the regulatory subunits of heterotrimeric SnRK1 complex.
There are several notable differences in the composition of SnRK2 in Arabidopsis compared to other plants. The subclass III-type SnRK2s are the most ancestral SnRK2 kinases that can be found in semiterrestrial algae [32][54]. The moss Physcomitrella patens genome encodes only four SnRK2 genes (PpSnRK2A/2B/2C/2D), all of which are categorized into subclass III [55]. The Ppsnrk2a/b/c/d quadruple knockout mutant displayed drastic ABA insensitivity and reduced osmotic stress tolerance, indicating that the roles of subclass III SnRK2s in abiotic stress signaling are evolutionarily conserved between bryophytes and angiosperms [54].
2.2. Role of Arabidopsis Subclass I and II SnRK2s in Regulating Abiotic Stress Responses
In Arabidopsis, there are two subclass II SnRK2s (SRK2C/SnRK2.8 and SRK2F/SnRK2.7), both of which are strongly activated in response to osmotic stress but weakly to ABA [12] (Figure 1). The first reported SnRK2, PKABA1 from wheat, belongs to this subclass [13][34]. Overexpression of SRK2C confers drought tolerance in Arabidopsis transgenic plants [56]. However, by contrast to the severe phenotype observed in subclass III mutant (srk2dei), the Arabidopsis double mutant (srk2cf) did not showed remarkable phenotypic changes even under osmotic stress conditions [33], indicating only minor contributions to ABA and osmostress signaling. bZIP-type transcription factors, such as ABF3 and ENHANCED EM LEVEL (EEL), are suggested to be as putative SRK2C substrates in vitro [33]. On the other hand, SRK2C was shown to function in metabolic processes, suggesting its crucial roles in plant growth [57]. Given that subclass II-type SnRK2 has been acquired in lycophytes (e.g., Selaginella tamariscina), it could be an intermediate molecule during the transition from subclass III SnRK2 in algae to subclass I SnRK2 in seed plants [33].
Subclass I-type SnRK2s are found in most angiosperms, but not in bryophytes or algae. There are five members in the Arabidopsis genome (SRK2A/SnRK2.4, SRK2B/SnRK2.10, SRK2G/SnRK2.1, SRK2H/SnRK2.5 and SRK2J/SnRK2.9), all of which except for SRK2J are rapidly activated by osmotic stress perception prior to ABA accumulation [11][12] (Figure 1). Unlike subclass II and III SnRK2s, subclass I SnRK2s are not activated by ABA [11][12] (Figure 2). Although less is known about subclass I SnRK2s compared to subclass III SnRK2s, recent studies have shown that this clade of SnRK2 is also essential for plant growth and survival under water-deficit conditions. For example, under salt stress conditions, SRK2A and its homolog SRK2B are involved in the maintenance of root system architecture [58] and in the modulation of ROS homeostasis [59]. Additionally, under osmotic stress conditions, SRK2B phosphorylates two of the LATE EMBRYOGENESIS ABUNDANT (LEA) dehydrin proteins, EARLY RESPONSE TO DEHYDRATION 10 (ERD10) and ERD14 [60]. Phosphorylation of ERD14 by SRK2B are involved in the translocation of ERD14 from cytosol to nucleus [60]. Importantly, after osmotic stress perception, subclass I-type SnRK2s, such as SRK2A and SRK2G, translocate to cytosolic punctate structures, which is known as processing bodies (P-bodies) [58][61]. In P-bodies, subclass I SnRK2s interact with and phosphorylate the mRNA decapping activator VARICOSE (VCS) [61] (Figure 2). VCS is a component of the mRNA decapping complex, which mediates the removal of the mRNA 5′-m7G-cap, leading to exonuclease-mediated mRNA decay [62]. Both the quadruple knockout mutant of subclass I SnRK2s (srk2abgh) and artificial micro RNA (amiRNA)-mediated VCS-knockdown plants shows similar growth retardation under osmotic stress conditions, suggesting the common phenotype is due to misregulation of mRNA metabolism in response to osmotic stress [61]. Given the fact that ABA-responsive SnRK2s (subclass II and III) showed no interaction with VCS [61], subclass I SnRK2s could predominantly regulate VCS-mediated mRNA decay in early osmotic stress response. In addition, cross-species complementation attempts demonstrated that Arabidopsis subclass I SnRK2 could not complement the osmotic stress tolerance-related phenotype of the P. patens subclass III SnRK2s quadruple mutant (Ppsnrk2a/b/c/d), indicating that the functions of subclass I SnRK2s are not compatible with subclass III SnRK2s [54]. In conclusion, recent investigations proposed that subclass I SnRK2s could be involved in plant growth and osmotic stress tolerance in a different manner from subclass II/ III SnRK2s, but both subclass I SnRK2-mediated and subclass II/ III SnRK2-mediated osmostress signaling are vital for plant to adopt to unfavorable conditions (Figure 2).
2.3. Other Regulators of Abiotic Stress Responses
In addition to the SnRK2 family, other protein kinases such as RECEPTOR-LIKE KINASES (RLKs), MITOGEN-ACTIVATED PROTEIN KINASES (MAPKs), CALCIUM-DEPENDENT PROTEIN KINASES (CDPKs/CPKs) contribute to plant responses to various abiotic stresses. For example, a plasma membrane localized receptor-like kinase GUARD CELL HYDROGEN PEROXIDE-RESISTANT 1 (GHR1) phosphorylates SLAC1 and is involved in both ABA- and H2O2-mediated stomatal closure [63]. Another transmembrane receptor-like kinase FERONIA (FER) and its ligand RAPID ALKALINIZATION FACTOR 1 (RALF1) interact with and activate the GTPase RHO-RELATED PROTEIN FROM PLANTS 11 (ROP11), thereby activating PP2C ABI2 to suppress ABA signaling [64]. RALF1-FER complex-dependent phosphorylation of GLYCINE-RICH RNA BINDING PROTEIN 7 (GRP7) affects alternative splicing to modulate stress responses and growth [65]. MPK9 and MPK12 are highly and preferentially expressed in guard cells and are involved in both ABA- and H2O2-mediated stomatal closure [66]. ABA-inducible MAPKKK17/18 activates MAPKK3–MPK1/2/7/14 cascade to regulate ABA-mediated inhibition of seed germination and cotyledon greening [67]. MAPKKK20, also known as ABA-INSENSITIVE PROTEIN KINASE 1 (AIK1), activates MAPKK5–MPK6 cascade to promote ABA-mediated primary root growth inhibition and stomatal closure and the PP2C ABI1 restricts AIK1 activity by dephosphorylation [68]. CPKs also mediate abiotic stress signaling through phosphorylation of substrate proteins, some of which are partially overlapped with SnRK2s. One of the examples is CPK6, which directly phosphorylates the N-terminal region of SLAC1 at Ser-59 and activates SLAC1 to promote stomatal closure [69]. Notably, transgenic plants carrying a SLAC1 transgene with mutations that produce a single substitution of Ser-59 to Ala (S59A), or the OST1-target site Ser-120 to Ala (S120A), rescued the slac1 knockout mutant phenotype. However, a transgenic line carrying a SLAC1 transgene with mutations that produce both Ser to Ala substitutions did not, suggesting that both CPK6- and OST1-mediated phosphorylation is essential for SLAC1 activation [69]. In addition to those, several protein kinases are modulating plant responses to abiotic stresses, which have been extensively discussed in the excellent recent reviews [70][71].
References
- Champion, A.; Kreis, M.; Mockaitis, K.; Picaud, A.; Henry, Y. Arabidopsis Kinome: After the Casting. Funct. Integr. Genom. 2004, 4, 163–187.
- Van Wijk, K.J.; Friso, G.; Walther, D.; Schulze, W.X. Meta-Analysis of Arabidopsis Thaliana Phospho-Proteomics Data Reveals Compartmentalization of Phosphorylation Motifs. Plant Cell 2014, 26, 2367–2389.
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Lo Muzio, L. The Crucial Role of Protein Phosphorylation in Cell Signaling and Its Use as Targeted Therapy (Review). Int. J. Mol. Med. 2017, 40, 271–280.
- Bhaskara, G.B.; Wong, M.M.; Verslues, P.E. The Flip Side of Phospho-Signalling: Regulation of Protein Dephosphorylation and the Protein Phosphatase 2Cs. Plant Cell Environ. 2019, 42, 2913–2930.
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The ArPlant.abidopsis Information Resource (TAIR): Improved Gene Annotation and New Tools. Nucleic Acids Res. 2012, 40, D1202–D1210.
- Smoly, I.; Shemesh, N.; Ziv-Ukelson, M.; Ben-Zvi, A.; Yeger-Lotem, E. An Asymmetrically Balanced Organization of Kinases versus Phosphatases across Eukaryotes Determines Their Distinct Impacts. PLoS Comput. Biol. 2017, 13, e1005221.
- Umezawa, T.; Nakashima, K.; Miyakawa, T.; Kuromori, T.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular Basis of the Core Regulatory Network in ABA Responses: Sensing, Signaling and Transport. Plant Cell Physiol. 2010, 51, 1821–1839.
- Mao, X.; Li, Y.; Rehman, S.U.; Miao, L.; Zhang, Y.; Chen, X.; Yu, C.; Wang, J.; Li, C.; Jing, R. The Sucrose Non-Fermenting 1-Related Protein Kinase 2 (SnRK2) Genes Are Multifaceted Players in Plant Growth, Development and Response to Environmental Stimuli. Plant Cell Physiol. 2020, 61, 225–242.
- Finkelstein, R. Abscisic Acid Synthesis and Response. Arab. Book 2013, 11.
- Shinozaki, K.; Yamaguchi-Shinozaki, K.; Seki, M. Regulatory Network of Gene Expression in the Drought and Cold Stress Responses. Curr. Opin. Plant Biol. 2003, 6, 410–417.
- Yoshida, R.; Umezawa, T.; Mizoguchi, T.; Takahashi, S.; Takahashi, F.; Shinozaki, K. The Regulatory Domain of SRK2E/OST1/SnRK2.6 Interacts with ABI1 and Integrates Abscisic Acid (ABA) and Osmotic Stress Signals Controlling Stomatal Closure in Arabidopsis. J. Biol. Chem. 2006, 281, 5310–5318.
- Boudsocq, M.; Barbier-Brygoo, H.; Laurière, C. Identification of Nine Sucrose Nonfermenting 1-Related Protein Kinases 2 Activated by Hyperosmotic and Saline Stresses in Arabidopsis Thaliana. J. Biol. Chem. 2004, 279, 41758–41766.
- Kobayashi, Y.; Yamamoto, S.; Minami, H.; Kagaya, Y.; Hattori, T. Differential Activation of the Rice Sucrose Nonfermenting1–Related Protein Kinase2 Family by Hyperosmotic Stress and Abscisic Acid. Plant Cell 2004, 16, 1163–1177.
- Umezawa, T.; Sugiyama, N.; Takahashi, F.; Anderson, J.C.; Ishihama, Y.; Peck, S.C.; Shinozaki, K. Genetics and Phosphoproteomics Reveal a Protein Phosphorylation Network in the Abscisic Acid Signaling Pathway in Arabidopsis Thaliana. Sci. Signal. 2013, 6, rs8.
- Wang, P.; Xue, L.; Batelli, G.; Lee, S.; Hou, Y.-J.; Oosten, M.J.V.; Zhang, H.; Tao, W.A.; Zhu, J.-K. Quantitative Phosphoproteomics Identifies SnRK2 Protein Kinase Substrates and Reveals the Effectors of Abscisic Acid Action. Proc. Natl. Acad. Sci. USA 2013, 110, 11205–11210.
- Park, S.-Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.; et al. Abscisic Acid Inhibits PP2Cs via the PYR/PYL Family of ABA-Binding START Proteins. Science 2009, 324, 1068–1071.
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C Phosphatase Activity Function as Abscisic Acid Sensors. Science 2009.
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Ishihama, Y.; Hirayama, T.; Shinozaki, K. Type 2C Protein Phosphatases Directly Regulate Abscisic Acid-Activated Protein Kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593.
- Vlad, F.; Rubio, S.; Rodrigues, A.; Sirichandra, C.; Belin, C.; Robert, N.; Leung, J.; Rodriguez, P.L.; Lauriere, C.; Merlot, S. Protein Phosphatases 2C Regulate the Activation of the Snf1-Related Kinase OST1 by Abscisic Acid in Arabidopsis. Plant Cell 2009, 21, 3170–3184.
- Saruhashi, M.; Ghosh, T.K.; Arai, K.; Ishizaki, Y.; Hagiwara, K.; Komatsu, K.; Shiwa, Y.; Izumikawa, K.; Yoshikawa, H.; Umezawa, T.; et al. Plant Raf-like Kinase Integrates Abscisic Acid and Hyperosmotic Stress Signaling Upstream of SNF1-Related Protein Kinase2. Proc. Natl. Acad. Sci. USA 2015, 112, E6388–E6396.
- Katsuta, S.; Masuda, G.; Bak, H.; Shinozawa, A.; Kamiyama, Y.; Umezawa, T.; Takezawa, D.; Yotsui, I.; Taji, T.; Sakata, Y. Arabidopsis Raf-like Kinases Act as Positive Regulators of Subclass III SnRK2 in Osmostress Signaling. Plant J. 2020, 103, 634–644.
- Takahashi, Y.; Zhang, J.; Hsu, P.-K.; Ceciliato, P.H.O.; Zhang, L.; Dubeaux, G.; Munemasa, S.; Ge, C.; Zhao, Y.; Hauser, F.; et al. MAP3Kinase-Dependent SnRK2-Kinase Activation Is Required for Abscisic Acid Signal Transduction and Rapid Osmotic Stress Response. Nat. Commun. 2020, 11, 12.
- Soma, F.; Takahashi, F.; Suzuki, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Plant Raf-like Kinases Regulate the MRNA Population Upstream of ABA-Unresponsive SnRK2 Kinases under Drought Stress. Nat. Commun. 2020, 11, 1–12.
- Lin, Z.; Li, Y.; Zhang, Z.; Liu, X.; Hsu, C.-C.; Du, Y.; Sang, T.; Zhu, C.; Wang, Y.; Satheesh, V.; et al. A RAF-SnRK2 Kinase Cascade Mediates Early Osmotic Stress Signaling in Higher Plants. Nat. Commun. 2020, 11, 1–10.
- Wang, P.; Zhao, Y.; Li, Z.; Hsu, C.-C.; Liu, X.; Fu, L.; Hou, Y.-J.; Du, Y.; Xie, S.; Zhang, C.; et al. Reciprocal Regulation of the TOR Kinase and ABA Receptor Balances Plant Growth and Stress Response. Mol. Cell 2018, 69, 100–112.e6.
- Belda-Palazón, B.; Adamo, M.; Valerio, C.; Ferreira, L.J.; Confraria, A.; Reis-Barata, D.; Rodrigues, A.; Meyer, C.; Rodriguez, P.L.; Baena-González, E. A Dual Function of SnRK2 Kinases in the Regulation of SnRK1 and Plant Growth. Nat. Plants 2020, 6, 1345–1353.
- Fu, L.; Wang, P.; Xiong, Y. Target of Rapamycin Signaling in Plant Stress Responses. Plant Physiol. 2020, 182, 1613–1623.
- Hrabak, E.M.; Chan, C.W.M.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.; Kudla, J.; Luan, S.; Nimmo, H.G.; Sussman, M.R.; et al. The Arabidopsis CDPK-SnRK Superfamily of Protein Kinases. Plant Physiol. 2003, 132, 666–680.
- Broeckx, T.; Hulsmans, S.; Rolland, F. The Plant Energy Sensor: Evolutionary Conservation and Divergence of SnRK1 Structure, Regulation, and Function. J. Exp. Bot. 2016, 67, 6215–6252.
- Zhu, J.-K.; Liu, J.; Xiong, L. Genetic Analysis of Salt Tolerance in Arabidopsis: Evidence for a Critical Role of Potassium Nutrition. Plant Cell 1998, 10, 1181–1191.
- Colina, F.; Amaral, J.; Carbó, M.; Pinto, G.; Soares, A.; Cañal, M.J.; Valledor, L. Genome-Wide Identification and Characterization of CKIN/SnRK Gene Family in Chlamydomonas Reinhardtii. Sci. Rep. 2019, 9, 350.
- Lind, C.; Dreyer, I.; López-Sanjurjo, E.J.; von Meyer, K.; Ishizaki, K.; Kohchi, T.; Lang, D.; Zhao, Y.; Kreuzer, I.; Al-Rasheid, K.A.S.; et al. Stomatal Guard Cells Co-Opted an Ancient ABA-Dependent Desiccation Survival System to Regulate Stomatal Closure. Curr. Biol. 2015, 25, 928–935.
- Mizoguchi, M.; Umezawa, T.; Nakashima, K.; Kidokoro, S.; Takasaki, H.; Fujita, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two Closely Related Subclass II SnRK2 Protein Kinases Cooperatively Regulate Drought-Inducible Gene Expression. Plant Cell Physiol. 2010, 51, 842–847.
- Anderberg, R.J.; Walker-Simmons, M.K. Isolation of a Wheat CDNA Clone for an Abscisic Acid-Inducible Transcript with Homology to Protein Kinases. Proc. Natl. Acad. Sci. USA 1992, 89, 10183–10187.
- Li, J.; Wang, X.-Q.; Watson, M.B.; Assmann, S.M. Regulation of Abscisic Acid–Induced Stomatal Closure and Anion Channels by Guard Cell AAPK Kinase. Science 2000, 287, 300–303.
- Yoshida, R.; Hobo, T.; Ichimura, K.; Mizoguchi, T.; Takahashi, F.; Aronso, J.; Ecker, J.R.; Shinozaki, K. ABA-Activated SnRK2 Protein Kinase Is Required for Dehydration Stress Signaling in Arabidopsis. Plant Cell Physiol. 2002, 43, 1473–1483.
- Mustilli, A.-C.; Merlot, S.; Vavasseur, A.; Fenzi, F.; Giraudat, J. Arabidopsis OST1 Protein Kinase Mediates the Regulation of Stomatal Aperture by Abscisic Acid and Acts Upstream of Reactive Oxygen Species Production. Plant Cell 2002, 14, 3089–3099.
- Kulik, A.; Wawer, I.; Krzywińska, E.; Bucholc, M.; Dobrowolska, G. SnRK2 Protein Kinases—Key Regulators of Plant Response to Abiotic Stresses. OMICS 2011, 15, 859–872.
- Belin, C.; de Franco, P.-O.; Bourbousse, C.; Chaignepain, S.; Schmitter, J.-M.; Vavasseur, A.; Giraudat, J.; Barbier-Brygoo, H.; Thomine, S. Identification of Features Regulating OST1 Kinase Activity and OST1 Function in Guard Cells. Plant Physiol. 2006, 141, 1316–1327.
- Boudsocq, M.; Droillard, M.-J.; Barbier-Brygoo, H.; Laurière, C. Different Phosphorylation Mechanisms Are Involved in the Activation of Sucrose Non-Fermenting 1 Related Protein Kinases 2 by Osmotic Stresses and Abscisic Acid. Plant Mol. Biol. 2007, 63, 491–503.
- Fujii, H.; Verslues, P.E.; Zhu, J.-K. Identification of Two Protein Kinases Required for Abscisic Acid Regulation of Seed Germination, Root Growth, and Gene Expression in Arabidopsis. Plant Cell 2007, 19, 485–494.
- Fujii, H.; Zhu, J.-K. Arabidopsis Mutant Deficient in 3 Abscisic Acid-Activated Protein Kinases Reveals Critical Roles in Growth, Reproduction, and Stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385.
- Fujita, Y.; Nakashima, K.; Yoshida, T.; Katagiri, T.; Kidokoro, S.; Kanamori, N.; Umezawa, T.; Fujita, M.; Maruyama, K.; Ishiyama, K.; et al. Three SnRK2 Protein Kinases Are the Main Positive Regulators of Abscisic Acid Signaling in Response to Water Stress in Arabidopsis. Plant Cell Physiol. 2009, 50, 2123–2132.
- Nakashima, K.; Fujita, Y.; Kanamori, N.; Katagiri, T.; Umezawa, T.; Kidokoro, S.; Maruyama, K.; Yoshida, T.; Ishiyama, K.; Kobayashi, M.; et al. Three Arabidopsis SnRK2 Protein Kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, Involved in ABA Signaling Are Essential for the Control of Seed Development and Dormancy. Plant Cell Physiol. 2009, 50, 1345–1363.
- Furihata, T.; Maruyama, K.; Fujita, Y.; Umezawa, T.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Abscisic Acid-Dependent Multisite Phosphorylation Regulates the Activity of a Transcription Activator AREB1. Proc. Natl. Acad. Sci. USA 2006, 103, 1988–1993.
- Yoshida, T.; Fujita, Y.; Maruyama, K.; Mogami, J.; Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Four Arabidopsis AREB/ABF Transcription Factors Function Predominantly in Gene Expression Downstream of SnRK2 Kinases in Abscisic Acid Signalling in Response to Osmotic Stress: Transcriptional Regulation in SnRK2 Signalling. Plant Cell Environ. 2015, 38, 35–49.
- Geiger, D.; Scherzer, S.; Mumm, P.; Stange, A.; Marten, I.; Bauer, H.; Ache, P.; Matschi, S.; Liese, A.; Al-Rasheid, K.A.S.; et al. Activity of Guard Cell Anion Channel SLAC1 Is Controlled by Drought-Stress Signaling Kinase-Phosphatase Pair. Proc. Natl. Acad. Sci. USA 2009, 106, 21425–21430.
- Lee, S.C.; Lan, W.; Buchanan, B.B.; Luan, S. A Protein Kinase-Phosphatase Pair Interacts with an Ion Channel to Regulate ABA Signaling in Plant Guard Cells. Proc. Natl. Acad. Sci. USA 2009, 106, 21419–21424.
- Imes, D.; Mumm, P.; Böhm, J.; Al-Rasheid, K.A.S.; Marten, I.; Geiger, D.; Hedrich, R. Open Stomata 1 (OST1) Kinase Controls R–Type Anion Channel QUAC1 in Arabidopsis Guard Cells. Plant J. 2013, 74, 372–382.
- Hosy, E.; Vavasseur, A.; Mouline, K.; Dreyer, I.; Gaymard, F.; Porée, F.; Boucherez, J.; Lebaudy, A.; Bouchez, D.; Véry, A.-A.; et al. The Arabidopsis Outward K+ Channel GORK Is Involved in Regulation of Stomatal Movements and Plant Transpiration. Proc. Natl. Acad. Sci. USA 2003, 100, 5549–5554.
- Sato, A.; Sato, Y.; Fukao, Y.; Fujiwara, M.; Umezawa, T.; Shinozaki, K.; Hibi, T.; Taniguchi, M.; Miyake, H.; Goto, D.B.; et al. Threonine at Position 306 of the KAT1 Potassium Channel Is Essential for Channel Activity and Is a Target Site for ABA-Activated SnRK2/OST1/SnRK2.6 Protein Kinase. Biochem. J. 2009, 424, 439–448.
- Sirichandra, C.; Gu, D.; Hu, H.-C.; Davanture, M.; Lee, S.; Djaoui, M.; Valot, B.; Zivy, M.; Leung, J.; Merlot, S.; et al. Phosphorylation of the Arabidopsis AtrbohF NADPH Oxidase by OST1 Protein Kinase. FEBS Lett. 2009, 583, 2982–2986.
- Yan, J.; Wang, P.; Wang, B.; Hsu, C.-C.; Tang, K.; Zhang, H.; Hou, Y.-J.; Zhao, Y.; Wang, Q.; Zhao, C.; et al. The SnRK2 Kinases Modulate MiRNA Accumulation in Arabidopsis. PLoS Genet. 2017, 13, e1006753.
- Shinozawa, A.; Otake, R.; Takezawa, D.; Umezawa, T.; Komatsu, K.; Tanaka, K.; Amagai, A.; Ishikawa, S.; Hara, Y.; Kamisugi, Y.; et al. SnRK2 Protein Kinases Represent an Ancient System in Plants for Adaptation to a Terrestrial Environment. Commun. Biol. 2019, 2, 30.
- Rensing, S.A.; Lang, D.; Zimmer, A.D.; Terry, A.; Salamov, A.; Shapiro, H.; Nishiyama, T.; Perroud, P.-F.; Lindquist, E.A.; Kamisugi, Y.; et al. The Physcomitrella Genome Reveals Evolutionary Insights into the Conquest of Land by Plants. Science 2008, 319, 64–69.
- Umezawa, T.; Yoshida, R.; Maruyama, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. SRK2C, a SNF1-Related Protein Kinase 2, Improves Drought Tolerance by Controlling Stress-Responsive Gene Expression in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2004, 101, 17306–17311.
- Shin, R.; Alvarez, S.; Burch, A.Y.; Jez, J.M.; Schachtman, D.P. Phosphoproteomic Identification of Targets of the Arabidopsis Sucrose Nonfermenting-like Kinase SnRK2.8 Reveals a Connection to Metabolic Processes. Proc. Natl. Acad. Sci. USA 2007, 104, 6460–6465.
- McLoughlin, F.; Galvan-Ampudia, C.S.; Julkowska, M.M.; Caarls, L.; van der Does, D.; Laurière, C.; Munnik, T.; Haring, M.A.; Testerink, C. The Snf1-Related Protein Kinases SnRK2.4 and SnRK2.10 Are Involved in Maintenance of Root System Architecture during Salt Stress. Plant J. 2012, 72, 436–449.
- Szymańska, K.P.; Polkowska-Kowalczyk, L.; Lichocka, M.; Maszkowska, J.; Dobrowolska, G. SNF1-Related Protein Kinases SnRK2.4 and SnRK2.10 Modulate ROS Homeostasis in Plant Response to Salt Stress. Int. J. Mol. Sci. 2019, 20, 143.
- Maszkowska, J.; Dębski, J.; Kulik, A.; Kistowski, M.; Bucholc, M.; Lichocka, M.; Klimecka, M.; Sztatelman, O.; Szymańska, K.P.; Dadlez, M.; et al. Phosphoproteomic Analysis Reveals That Dehydrins ERD10 and ERD14 Are Phosphorylated by SNF1-Related Protein Kinase 2.10 in Response to Osmotic Stress. Plant Cell Environ. 2019, 42, 931–946.
- Soma, F.; Mogami, J.; Yoshida, T.; Abekura, M.; Takahashi, F.; Kidokoro, S.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-Unresponsive SnRK2 Protein Kinases Regulate MRNA Decay under Osmotic Stress in Plants. Nat. Plants 2017, 3, 16204.
- Chantarachot, T.; Bailey-Serres, J. Polysomes, Stress Granules, and Processing Bodies: A Dynamic Triumvirate Controlling Cytoplasmic MRNA Fate and Function. Plant Physiol. 2018, 176, 254–269.
- Hua, D.; Wang, C.; He, J.; Liao, H.; Duan, Y.; Zhu, Z.; Guo, Y.; Chen, Z.; Gong, Z. A Plasma Membrane Receptor Kinase, GHR1, Mediates Abscisic Acid- and Hydrogen Peroxide-Regulated Stomatal Movement in Arabidopsis. Plant Cell 2012, 24, 2546–2561.
- Yu, F.; Qian, L.; Nibau, C.; Duan, Q.; Kita, D.; Levasseur, K.; Li, X.; Lu, C.; Li, H.; Hou, C.; et al. FERONIA Receptor Kinase Pathway Suppresses Abscisic Acid Signaling in Arabidopsis by Activating ABI2 Phosphatase. Proc. Natl. Acad. Sci. USA 2012, 109, 14693–14698.
- Wang, L.; Yang, T.; Wang, B.; Lin, Q.; Zhu, S.; Li, C.; Ma, Y.; Tang, J.; Xing, J.; Li, X.; et al. RALF1-FERONIA Complex Affects Splicing Dynamics to Modulate Stress Responses and Growth in Plants. Sci. Adv. 2020, 6, eaaz1622.
- Jammes, F.; Song, C.; Shin, D.; Munemasa, S.; Takeda, K.; Gu, D.; Cho, D.; Lee, S.; Giordo, R.; Sritubtim, S.; et al. MAP Kinases MPK9 and MPK12 Are Preferentially Expressed in Guard Cells and Positively Regulate ROS-Mediated ABA Signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 20520–20525.
- Danquah, A.; de Zélicourt, A.; Boudsocq, M.; Neubauer, J.; Frei dit Frey, N.; Leonhardt, N.; Pateyron, S.; Gwinner, F.; Tamby, J.-P.; Ortiz-Masia, D.; et al. Identification and Characterization of an ABA-Activated MAP Kinase Cascade in Arabidopsis Thaliana. Plant J. 2015, 82, 232–244.
- Li, K.; Yang, F.; Zhang, G.; Song, S.; Li, Y.; Ren, D.; Miao, Y.; Song, C.-P. AIK1, A Mitogen-Activated Protein Kinase, Modulates Abscisic Acid Responses through the MKK5-MPK6 Kinase Cascade. Plant Physiol. 2017, 173, 1391–1408.
- Brandt, B.; Munemasa, S.; Wang, C.; Nguyen, D.; Yong, T.; Yang, P.G.; Poretsky, E.; Belknap, T.F.; Waadt, R.; Alemán, F.; et al. Calcium Specificity Signaling Mechanisms in Abscisic Acid Signal Transduction in Arabidopsis Guard Cells. eLife 2015, 4, e03599.
- Chen, X.; Ding, Y.; Yang, Y.; Song, C.; Wang, B.; Yang, S.; Guo, Y.; Gong, Z. Protein Kinases in Plant Responses to Drought, Salt, and Cold Stress. J. Integr. Plant Biol. 2021, 63, 53–78.
- Zhang, H.; Zhao, Y.; Zhu, J.-K. Thriving under Stress: How Plants Balance Growth and the Stress Response. Dev. Cell 2020, 55, 529–543.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.4K
Revisions:
2 times
(View History)
Update Date:
26 Jul 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




