
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Yaqi Hao | + 3131 word(s) | 3131 | 2021-07-06 14:15:57 | | | |
| 2 | Beatrix Zheng | Meta information modification | 3131 | 2021-07-16 11:09:36 | | |
Video Upload Options
The basic helix-loop-helix (bHLH) transcription factor family is one of the largest transcription factor gene families in Arabidopsis thaliana, and contains a bHLH motif that is highly conserved throughout eukaryotic organisms. Members of this family have two conserved motifs, a basic DNA binding region and a helix-loop-helix (HLH) region. These proteins containing bHLH domain usually act as homo- or heterodimers to regulate the expression of their target genes, which are involved in many physiological processes and have a broad range of functions in biosynthesis, metabolism and transduction of plant hormones.
1. Introduction
2. Functions of bHLH Factors in Plant Growth and Development in Arabidopsis
Seed dormancy and germination are critical processes in the lifespan of plants, which are mediated by various external environmental factors such as temperature, light and humidity, and internal factors, such as phytohormones. Two type of phytohormones, ABA and gibberellin (GA), are crucial regulators of seed dormancy and germination. ABA enhances seed dormancy, whereas GA breaks dormancy and promotes germination. NCED (9-cis-epoxycarotenoid dioxygenase) is considered to catalyze the first dedicated step in ABA biosynthesis [22], ODR1 (reversal of rdo5, an ortholog of the rice seed dormancy 4
In addition to phytohormones, photoreceptors also regulate seed dormancy and germination. Phytochrome interacting factors (PIFs), a group of typical of bHLH proteins, could interact with diverse groups of factors to integrate external environmental and internal signals, and further control seed germination, shade avoidance and crosstalk of plant hormones and the clock-derived signaling pathway [23][24]. The phytochrome-interacting bHLH protein PIL5 (PIF3-like 5 PIF1/bHLH15) is a repressor of seed germination, acting by reducing GA level in the dark; phytochromes accelerate the PIL5 degradation and increase the level of bioactive GA in seeds upon exposure to light. In addition, PIL5 activates the expression of the ABA synthesis gene and maintains a high level of ABA in the dark to inhibit seed germination [25].
It is critical for plants to manage their flowering time, which is regulated by an intricate network of molecular signaling and controlled by various environmental factors such as photoperiod and temperature. The CONSTANS (CO) protein functions as an essential component for transforming biological clock signals into flowering signals to initiate plant flowering. Cryptochromes (CRY1/2) are blue light receptors that inhibit hypocotyl elongation and control floral initiation [26]. CRY2, CIB1 and CO can form a protein complex in response to blue light and then promote floral initiation [27][28].
Recently, a bHLH transcription factor, named as NO FLOWERING IN SHORT DAY (NFL) was shown to be necessary for the promotion of flowering specifically under short-day (SD) conditions, andnflmutants did not flower under SD conditions but were similar to wild type under long-day (LD) conditions [29]. Additionally, three myelocytomatosis (MYC) proteins redundantly regulate flowering under both LD and SD conditions. Moreover, the Brassinosteroid Enhanced Expression1 (BEE1, bHLH44) protein is stabilized under blue light; this protein is an integrator regulating photoperiodic flowering [30].
In Arabidopsis, temperature also affects the flowering time, and high temperature (29 °C) not only induces rapid hypocotyl elongation but also results in early flowering. In addition, the central integrator bHLH transcription factor phytochrome-interacting factor 4 (PIF4) can accelerate the flowering process by activating FT at high temperature conditions [31]. In addition to regulating the flowering time, bHLH family members are involved in the flower organ development. SPATULA (SPT) encodes a bHLH transcription factor and plays a role in floral morphogenesis processes as previously described [32].
In summary, the flowering time is strictly controlled by an intricate network, and bHLH family members act together with many other proteins to allow plants to flower at suitable time and under favorable environments.
Plants process a cell determination mechanism for formatting specific cell types, and this system relies on the expression of different genes in a proper spatiotemporal manner. Many transcription factors participate in this critical process, including a number of bHLH transcription factors, which play essential roles in the root and shoot cell fate determination [33]. During the root development, two types of cells arise from the epidermis: root hair cells and non-hair cells.
bHLH83 (root hair defective 6, RHD6), is a transcription factor that has fewer root hairs and functions in the inhibition root hair formation in cortical cells [13][33], and another bHLH protein, ROOT HAIR DEFECTIVE Additional genes, such asRSL3/4andLRL3(Lj-RHL1-LIKE3) also act as downstream of RHD6 and RSL1 to promote the root hair and cell differentiation [34][35][36]. GL2, a member of homeodomain-leucine zipper (HD-ZIP) protein, affects the epidermal cell fate including in trichomes, root hair and the seed coat. It represses the transcription of RHD6 to inhibit hair formation in N-cells (non-root hair cells), leading to the expression of non-hair genes [35][37].
Furthermore, ET is involved in root hair initiation and elongation, and ETHYLENE INSENSITIVE 3 (EIN3) and its homolog EIN3-like1 (EIL1) can coordinate with RHD6/RSL1 to upregulate RSL4 and then participate in root epidermis development [38]. Another bHLH complex named MYB-bHLH-WD40 was proposed to regulate the guard cell and root hair differentiation [39].
In addition to their roles in the root epidermis formation, the bHLH family is also involved in other cell fate related processes, including stamen and stomatal development. The IIIe bHLH transcription factor MYC5 (bHLH28) has redundant functions with MYC2 (bHLH6), MYC3 (bHLH5) and MYC4 (bHLH4), which interact with MYB21 and MYB24 to form the bHLH-MYB complex to regulate the stamen development [40]. In addition, FAMA (bHLH97) is specifically expressed in the stomata and has functions in halting proliferative division and promoting guard cells [41].
Overall, the establishment of this specification mechanism is complicated and also influenced by several plant growth regulators, including hormones such auxin (indole-3-acetic acid, IAA), ET, JA [42][38].
3. Functions in the Response to Light and Phytohormones
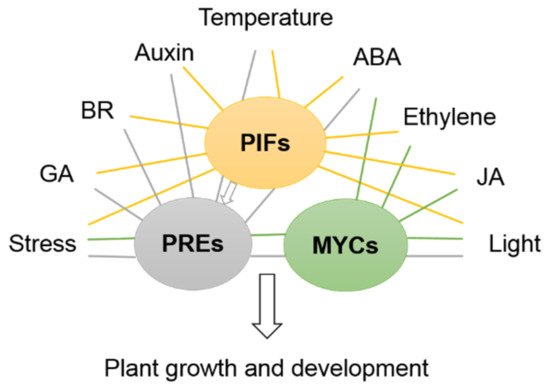
Under specific conditions, multiple phytohormones and environmental factors are in constant crosstalk with each other to affect the plant growth and development. Based on previous studies, GA, IAA, BR, ET and light are usually considered to promote cell expansion in plant growth, while ABA, JA and SA are normally involved in the response to biotic and abiotic stresses. bHLH proteins that respond to plant hormones and environmental factors participate in various processes. They are able to interact with each other to cooperatively or antagonistically modulate the plant growth.
Light is an important factor that influences the plant growth and development, and many bHLH transcription factors are reported to participate in this process by inducing the expression of related downstream genes and various light-mediated effects.
PIFs interact with the active form (Pfr) of phytochrome to modulate growth, including in response to environmental signals such as light and stress and via other signaling pathways [43]. The PIF family belongs to the bHLH superfamily VII of transcription factors, which play central roles in the regulation of light signaling. To date, 15 PIF members have already been identified, and 7 PIF members have been shown to bind the Pfr form of phyB in Arabidopsis, while other members do not interact with the light-activated phytochrome [13][14][44][45][46]. Each PIF has individualized or redundant biological functions with other PIF proteins during various responses.
In the dark, four PIFs (PIF1/PIL5, PIF3, PIF4 and PIF5/PIL6) directly interact with Pfr to promote skotomorphogenesis by repressing photomorphogenesis. Loss-of-function ofpifmutant showed phenotypes of reduced hypocotyl elongation. PIFs also interact with diverse groups of transcription factors to integrate external environmental and internal signals, including in seed germination, shade avoidance and crosstalk among plant hormones and clock-related signaling pathways [47][23]. A number of genes have been confirmed to be direct targets of PIF, thus mediating downstream light signaling networks through the PIFs.
PIF3 (bHLH8), the first identified member in the PIF family, acts as a regulator in the seedling de-etiolation and modulates both positive and negative response to phytochrome-mediated signaling [47][48]. PIF3 can also heterodimerize with the atypical basic bHLH protein HFR1 (long hypocotyl in far-red, bHLH26) to modulate phyA signaling [49]. In addition, PIF3 is involved in plant freezing response as a negatively regulator which can interact with EBF1 (EIN1-BINDING F-BOX1) through the CBF (C-REPEAT BINDING FACTOR) pathway [50].
PIF4 is a key transcription factor in the light signaling pathway; it interacts selectively with Pfr and negatively regulates phyB signaling in Arabidopsis [51], Phytochrome Rapidly Regulated 1 (PAR1, bHLH165) and its homolog PAR2 (bHLH166) lack the ability to directly interact with phytochromes that are rapidly induced by shade [52][53], while PAR1 directly interacts with PIF4 to form a heterodimer to inhibit PIF4 function in cell elongation [54]. Moreover, growing evidence indicates that PIF4 acts as central regulator, coordinating plant response to multiple environmental signals [55].
The PIL (PIF3-like proteins, PILs) (bHLH15) is a component that negatively regulate the chlorophyll biosynthetic pathway, seeds germination and inhibit hypocotyl elongation in the dark. The activity of PIF1 was repressed by phyA and phyB in light, and regulated ABA signaling [56][57][58][59]. In addition, other PIFs, such as PIF5 (PIL6), PIF6 (PIL2) and PIF7, have been shown to interact with the Pfr form of phyB or phyA, which are involved in phytochrome signaling [60][61][62].
Taken together, the bHLH proteins PILs/PIFs are proposed to form heterodimers to regulate bHLH network activity and are central components that integrate multiple signals in response to light.
The phytohormone jasmonate acid (JA) plays a vital role in the plant development and the response to various stresses. The presence of JA triggers the key protein jasmonate ZIM-domain (JAZ) to interact with Coronatine Insensitive 1 (COI1), part of the SCFCOI1ubiquitin E3 ligase complex. Then, JAZ proteins were degraded by the 26S protease, resulting in multiple transcription factors free from JAZ-mediated repression and further activating downstream JA-mediated responsive genes. The homologs of MYC2, MYC3 (bHLH5) and MYC4 (bHLH4) are known to form homodimers/heterodimers, and they can also bind to the G-box involved in the JA signaling pathway but exhibit gene redundancy with MYC2 [63][64].
The bHLH transcription factor GLABRA3 (GL3, bHLH1) can form a WD-repeat/bHLH/MYB complex with TRANSPARENT TESTA (TTG1) and the R2R3-MYB transcription factor GLABRA1 (GL1), which is repressed by JAZs and DELLAs, is responsible for trichome initiation [65][66][67][68].
Four subgroup IVa bHLH transcription factors (bHLH18, bHLH19, bHLH20 and bHLH25) can be induced by JA and inhibit the transcription of the FIT and Ib bHLH genes, which have been suggested to function redundantly in JA-mediated FIT protein degradation in the presence of JA or under iron deficient conditions via the 26S proteasome pathway [69].
The jasmonate-activated transcription factor MYC2 has also been found to interact with the key component in the ET signaling pathway EIN3 and its close homolog EIL1 to repressed its DNA binding activity and affect hook formation [70], suggesting that jasmonate and ET have antagonistic functions during apical hook development.
The plant hormone IAA has multiple roles in the plant growth and development, such as in cell division, cell elongation and cell differentiation, which are affected by the regulation of IAA response genes [71]. BIGPETALp (BPEp, bHLH31) is a bHLH transcription factor that can interact with AUXIN RESPONSE FACTOR8 (ARF8) to influence cell expansion and petal growth [72], PRE6 is a target of ARF5 and ARF8 that negatively regulates auxin response genes in Arabidopsis [73].
Plant are constantly under extrinsic abiotic/biotic environmental stresses, including cold, drought, high salinity, pathogen and extreme temperature. In response to these diverse stresses, plants have evolved sophisticated adaptation mechanisms. To date, several bHLH transcription factors have been reported to mediate abiotic and biotic stress signaling pathways to regulate plant responses in Arabidopsis in different ways.
The plant hormone ABA plays a central role in a variety of physiological processes and environmental response involved in plant growth, including responses to drought, cold, heat and salinity stresses [74][75][76]. Several bHLH transcription factors, such as bHLH112, MYC2, AIB, AtAIG1, bHLH129 and bHLH92, have been reported to be involved in the regulation of ABA signaling via these processes [77][78][79][80].
A loss-of-function mutant of bHLH112, a transcriptional activator, displayed a late-flowering phenotype under long day conditions in Arabidopsis, and its transcript level was correlated with salt and drought tolerance. bHLH112 regulates gene expression by binding to E-box and GCG-box motifs in the gene promoters and then mediate multiple physiological response to improve stress tolerance [77][81].
ABA-inducible bHLH-type transcription factor (AIB) and ABA-inducible gene (AIG1) are ABA-induced genes, the proteins contain a bHLH type DNA binding domain and play a positive role in ABA signaling in Arabidopsis [78][79]. MYC2 and MYB2 have been shown to be transcription activators that function in the ABA signal transduction pathway by directly regulating the expression of ABA response genes [17]. In addition, MYC2 can also be involved in JA [82][83][84].
The expression level of bHLH129 is reduced upon exogenously application of ABA, and bHLH129 regulates the expression of several ABA signaling component genes [80] to promote root elongation.
PREs are involved in plant growth and development and are also involved in the regulation of ABA mediated salt responses in Arabidopsis. Some PREs are ABA responsive genes; their expression levels are decreased under ABA treatment, and this response has functions in regulating plant growth and environmental stimuli [85].
ABA induces stomatal closure and changes the expression of numerous genes to adapt to drought stress, ABA-responsive kinase substrate (AKS1), also a bHLH transcription activator, is inhibited by ABA through phosphorylated to form monomer by SNF1-related protein kinase 2 (SnRK2) in Arabidopsis guard cells [86][87].
In addition, the BEE transcription factor family members (BEE1/2/3) regulate plant responses to abiotic stress. BEE genes are strongly repressed by ABA and are redundant negative regulators of physiological responses to abiotic stress, whereas the BEE2 dimerized protein IBH1 is a positive modulator that improves salt and drought tolerance [88]. NaCl-induced expression of bHLH92 confers tolerance to salt and osmotic stress which is partially dependent on ABA and SALT OVERLY SENSITIVE 2 (SOS2) [89].
Some bHLH transcription factors are involved in signal transduction networks mediated by plant hormones. Moreover, many bHLH transcription factors have been reported to be involved in the regulation of BRs, ABA and IAA signaling pathways. PREs participate in various hormone-, temperature- and light-responsive signaling pathways to regulate plant growth and development in many ways [90][91][92][93]. PRE2 (bHLH134) and PRE6 (bHLH163) are ABA response genes that affect plant sensitivity to ABA, indicating that some PREs are involved in ABA and salts responses [73].
Interacting Factors (AIFs), negatively regulate cell elongation in Arabidopsis [92][94]. Activators for cell elongation (ACEs) can promote cell elongation, while IBH1 interacts with ACEs to inhibit their functions in the induction of cell elongation. Another bHLH protein, homolog of BEE2 interacting with IBH1 (HBI1), is involved in BR-mediated growth to promote cell elongation [90]. Functional analysis showed that bHLH proteins have related functions but with different mechanisms to regulate cell elongation.
BEE1 (bHLH44), BEE2 (bHLH58) and BEE3 (bHLH50) are functionally redundant bHLH transcription factors for which expression is induced by BL treatment, indicating that they function in the early response to BRs and their expression is required by ABA [18]. In brief, the BEE1 protein is stabilized under blue light, which is an integrator to regulate photoperiodic flowering [95]. BEE2 and CIB1 negatively regulate immunity and are functionally redundant with HBI1 in plant immunity [90][96]. Moreover, BEEs positive regulate shade avoidance syndrome with BES1-INTERACTING MYC-LIKE protein (BIMs) [97][98].
4. Conclusions
References
- Murre, C.; McCaw, P.S.; Baltimore, D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 1989, 56, 777–783.
- Massari, M.E.; Murre, C. Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 2000, 20, 429–440.
- Duek, P.D.; Fankhauser, C. bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci. 2005, 10, 51–54.
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116.
- Atchley, W.R.; Fitch, W.M. A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl. Acad. Sci. USA 1997, 94, 5172–5176.
- Henriksson, M.; Lüscher, B. Proteins of the Myc network: Essential regulators of cell growth and differentiation. Adv. Cancer Res. 1996, 68, 109–182.
- Goding, C.R. Motif from neural crest to melanoma: Signal transduction and transcription in the melanocyte lineage. Genes Dev. 2000, 14, 1712–1728.
- Sun, X.H.; Copeland, N.G.; Jenkins, N.A.; Baltimore, D. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol. Cell. Biol. 1991, 11, 5603–5611.
- Ledent, V.; Vervoort, M. The basic helix-loop-helix protein family: Comparative genomics and phylogenetic analysis. Genome Res. 2001, 11, 754–770.
- Fisher, A.; Caudy, M. The function of hairy-related bHLH repressor proteins in cell fate decisions. Bioessays 1998, 20, 298–306.
- Crozatier, M.; Valle, D.; Dubois, L.; Ibnsouda, S.; Vincent, A. Collier, a novel regulator of Drosophila head development, is expressed in a single mitotic domain. Curr. Biol. 1996, 6, 707–718.
- Bailey, P.C.; Martin, C.; Toledo-Ortiz, G.; Quail, P.H.; Huq, E.; Heim, M.A.; Jakoby, M.; Werber, M.; Weisshaar, B. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 2003, 15, 2497–2501.
- Heim, M.A.; Jakoby, M.; Werber, M.; Marti, C.; Weisshaar, B.; Bailey, P.C. The basic Helix-Loop-Helix transcription factor family in plants: A genome-wide study of Protein structure and functional diversity. Mol. Biol. Evol. 2003, 20, 735–747.
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770.
- Buck, M.J.; Atchley, W.R. Phylogenetic Analysis of Plant Basic Helix-Loop-Helix Proteins. J. Mol. Evol. 2003, 56, 742–750.
- Castelain, M.; Hir, R.L.; Bellini, C. The non-DNA-binding bHLH transcription factor PRE3/bHLH135/ATBS1/TMO7 is involved in the regulation of light signaling pathway in Arabidopsis. Physiologia Plantarum 2012, 145, 450–460.
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78.
- Friedrichsen, D.M.; Nemhauser, J.; Muramitsu, T.; Maloof, J.N.; Alonso, J.; Ecker, J.R.; Furuya, M.; Chory, J. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 2002, 162, 1445–1456.
- Liu, Y.; Li, X.; Li, K.; Liu, H.; Lin, C. Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time in Arabidopsis. PLoS Genet. 2013, 9, e1003861.
- Yao, X.; Cai, Y.; Yu, D.; Liang, G. bHLH104 confers tolerance to cadmium stress in Arabidopsis thaliana. J. Integr. Plant Biol. 2018, 60, 691–702.
- Pires, N.; Dolan, L. Origin and diversification of Basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 2010, 27, 862–874.
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185.
- Ni, M.; Tepperman, J.M.; Quail, P.H. PIF3, a Phytochrome-interacting factor necessary for normal photoinduced signal transduction, as a novel basic helix-loop-helix protein. Cell 1998, 95, 657–667.
- Oh, E.; Zhu, J.; Wang, Z. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012, 14, 802–809.
- Oh, E.; Yamaguchi, S.; Kamiya, Y.; Bae, G.; Chung, W.I.; Choi, G. Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 2006, 47, 124–139.
- Guo, H.; Yang, H.; Mockler, T.; Lin, C. Regulation of flowering time by Arabidopsis photoreceptors. Science 1998, 279, 1360–1363.
- Liu, H.; Yu, X.; Li, K.; Klejnot, J.; Yang, H.; Lisiero, D.; Lin, C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 2008, 322, 1535–1539.
- Liu, Y.; Li, Y.; Ma, D.; Chen, Z.; Wang, J.; Liu, H. CIB1 and CO interact to mediate CRY2-dependent regulation of flowering. EMBO Rep. 2018, 19, e45762.
- Sharma, N.; Xin, R.; Kim, D.; Sung, S.; Lange, T.; Huq, E. NO FLOWERING IN SHORT DAY (NFL) is a bHLH transcription factor that promotes flowering specifically under short-day conditions in Arabidopsis. Development 2016, 143, 682–690.
- Wang, Z.; Yang, Z.; Li, F. Updates on molecular mechanisms in the development of branched trichome in Arabidopsis and nonbranched in cotton. Plant Biotechnol. J. 2019, 17, 1706–1722.
- Kumar, S.V.; Lucyshyn, D.; Jaeger, K.E.; Alós, E.; Alvey, E.; Harberd, N.P.; Wigge, P.A. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 2012, 484, 242–245.
- Heisler, M.G.; Atkinson, A.; Bylstra, Y.H.; Walsh, R.; Smyth, D.R. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 2001, 128, 1089–1098.
- Zhao, H.; Li, X.; Ma, L. Basic helix-loop-helix transcription factors and epidermal cell fate determination in Arabidopsis. Plant Signal. Behav. 2012, 7, 1556–1560.
- Yi, K.; Menand, B.; Bell, E.; Dolan, L. A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat. Genet. 2010, 42, 264–267.
- Bruex, A.; Kainkaryam, R.M.; Wieckowski, Y.; Kang, Y.H.; Bernhardt, C.; Xia, Y.; Zheng, X.; Wang, J.Y.; Lee, M.M.; Benfey, P.; et al. A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet. 2012, 8, e1002446.
- Han, X.; Zhang, M.; Yang, M.; Hu, Y. Arabidopsis JAZ proteins interact with and suppress RHD6 transcription factor to regulate jasmonate-stimulated root hair development. Plant Cell 2020, 32, 1049–1062.
- Masucci, J.D.; Rerie, W.G.; Foreman, D.R.; Zhang, M.; Galway, M.E.; Marks, M.D.; Schiefelbein, J.W. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 1996, 122, 1253–1260.
- Feng, Y.; Xu, P.; Li, B.; Li, P.; Wen, X.; An, F.; Gong, Y.; Xin, Y.; Zhu, Z.; Wang, Y.; et al. Ethylene promoters root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 13834–13839.
- Ramsay, N.A.; Glover, B.J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70.
- Qi, T.; Huang, H.; Song, S.; Xie, D. Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. Plant Cell 2015, 27, 1620–1633.
- Ohashi-Ito, K.; Bergmann, D.C. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 2006, 18, 2493–2505.
- Pitts, R.J.; Cernac, A.; Estelle, M. Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 1999, 16, 553–560.
- Paik, I.; Kathare, P.K.; Kim, J.; Huq, E. Expanding roles of PIFs in signal integration from multiple processes. Mol. Plant 2017, 10, 1035–1046.
- Castillon, A.; Shen, H.; Huq, E. Phytochrome Interacting Factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 2007, 12, 514–521.
- Leivar, P.; Quail, P.H. PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 2011, 16, 19–28.
- Leivar, P.; Monte, E. PIFs: Systems integrators in plant development. Plant Cell 2014, 26, 56–78.
- Liu, F.; Zhang, H.; Ding, L.; Soppe, W.J.J.; Xiang, Y. REVERSAL OF RDO5 1, a homolog of rice seed dormancy 4, interacts with bHLH57 and controls ABA biosynthesis and seed dormancy in Arabidopsis. Plant Cell 2020, 32, 1933–1948.
- Kim, J.; Yi, H.; Choi, G.; Shin, B.; Song, P.S.; Choi, G. Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 2003, 15, 2399–2407.
- Fairchild, C.D.; Schumaker, M.A.; Quail, P.H. HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 2000, 14, 2377–2391.
- Jiang, B.; Shi, Y.; Zhang, X.; Xin, X.; Qi, L.; Guo, H.; Li, J.; Yang, S. PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E6695–E6702.
- Huq, E.; Quail, P.H. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002, 21, 2441–2450.
- Roig-Villanova, I.; Bou, J.; Sorin, C.; Devlin, P.F.; Martínez-García, J.F. Identification of primary target genes of phytochrome signaling: Early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol. 2006, 141, 85–96.
- Roig-Villanova, I.; Bou-Torrent, J.; Galstyan, A.; Carretero-paulet, L.; Portolés, S.; Rodríguez-Concepción, M.; Martínez-García, J. Interaction of shade avoidance and auxin response: A role for two novel atypical bHLH proteins. EMBO J. 2007, 26, 4756–4767.
- Hao, Y.; Oh, E.; Choi, G.; Liang, Z.; Wang, Z.Y. Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant 2012, 5, 688–697.
- Brumbarova, T.; Ivanov, R. The nutrient response transcriptional regulome of Arabidopsis. Iscience 2019, 19, 358–368.
- Oh, E.; Kim, J.; Park, E.; Kim, J.; Kang, C.; Choi, G. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 2004, 16, 3045–3058.
- Oh, E.; Yamaguchi, S.; Hu, J.; Yusuke, J.; Jung, B.; Paik, I.; Lee, H.; Sun, T.; Kamiya, Y.; Choi, G. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis Seeds. Plant Cell 2007, 19, 1192–1208.
- Huq, E.; Al-Sady, B.; Hudson, M.; Kim, C.; Apel, K.; Quail, P.H. PHYTOCHROME-INTERACTING FACTOR 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 2004, 305, 1937–1941.
- Moon, J.; Zhu, L.; Shen, H.; Huq, E. PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc. Natl. Acad. Sci USA 2008, 105, 9433–9438.
- Shen, Y.; Khanna, R.; Carle, C.M.; Quail, P.H. Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 2007, 145, 1043–1051.
- Khanna, R.; Huq, E.; Kikis, E.A.; Al-Sady, B.; Lanzatella, C.; Quail, P.H. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 2004, 16, 3033–3044.
- Leivar, P.; Monte, E.; Al-Sady, B.; Carle, C.; Storer, A.; Alonso, J.M.; Ecker, J.R.; Quail, P.H. The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 2008, 20, 337–352.
- Fernández-Calvo, P.; Chini, A.; Fernández-Barbero, G.; Chico, J.M.; Gimenez-lbanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; José Manuel Franco-Zorrilla, J.M.; et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011, 23, 701–715.
- Schweizer, F.; Fernández-Calvo, P.; Zander, M.; Diez-Diaz, M.; Fonseca, S.; Glauser, G.; Lewsey, M.G.; Ecker, J.R.; Solano, R.; Reymond, P. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 2013, 25, 3117–3132.
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827.
- Zhao, M.; Morohashi, K.; Hatlestad, G.; Grotewold, E.; Lloyd, A. The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 2008, 135, 1991–1999.
- Qi, T.; Song, S.; Ren, Q.; Wu, D.; Huang, H.; Chen, Y.; Fan, M.; Peng, W.; Ren, C.; Xie, D. The Jasmonate-ZIM-Domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 2011, 23, 1795–1814.
- Qi, T.; Huang, H.; Wu, D.; Yan, J.; Qi, Y.; Song, S.; Xie, D. Arabidopsis DELLA and JAZ proteins bind the WD-Repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell 2014, 26, 1118–1133.
- Li, X.; Zhang, H.; Ai, Q.; Liang, G.; Yu, D. Two bHLH transcription factors, bHLH34 and bHLH104, regulate iron homeostasis in Arabidopsis thaliana. Plant Physiol. 2016, 170, 2478–2493.
- Zhang, X.; Zhu, Z.; An, F.; Hao, D.; Li, P.; Song, J.; Yi, C.; Guo, H. Jasmonate-activated MYC2 represses ETHYLENE INSENSITIVE3 activity to antagonize ethylene-promoted apical hook formation in Arabidopsis. Plant Cell 2014, 26, 1105–1117.
- Teale, W.D.; Paponov, I.A.; Palme, K. Auxin in action: Signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 2006, 7, 847–859.
- Varaud, E.; Brioudes, F.; Szécsi, J.; Leroux, J.; Brown, S.; Perrot-Rechenmann, C.; Bendahmane, M. AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell 2011, 23, 973–983.
- Zheng, K.; Wang, Y.; Zhang, N.; Jia, Q.; Wang, X.; Hou, C.; Chen, J.; Wang, S. Involvement of PACLOBUTRAZOL RESISTANCE6/KIDARI, an atypical bHLH transcription factor, in auxin responses in Arabidopsis. Front. Plant Sci. 2017, 8, 1813.
- Leung, J.; Giraudat, J. Abscisic acid signal transduction. Annu. Rev. Plant Phys. 1998, 49, 199–222.
- McCourt, P. Genetic analysis of hormone signaling. Annu Rev. Plant Phys. 1999, 50, 219–243.
- Finkelstein, R.R.; Gampala, S.S.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14, S15–S45.
- Liu, Y.; Ji, X.; Nie, X.; Qu, M.; Zheng, L.; Tan, Z.; Zhao, H.; Huo, L.; Liu, S.; Zhang, B.; et al. Arabidopsis AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs. N. Phytol. 2015, 207, 692–709.
- Kim, J.; Kim, H.Y. Molecular characterization of a bHLH transcription factor involved in Arabidopsis abscisic acid-mediated response. BBA Gene Struct. Expr. 2006, 1759, 191–194.
- Li, H.; Sun, J.; Xu, Y.; Jiang, H.; Wu, X.; Li, C. The bHLH-type transcription factor AtAIB positively regulates ABA response in Arabidopsis. Plant Mol. Biol. 2007, 65, 655–665.
- Tian, H.; Guo, H.; Dai, X.; Cheng, Y.; Zheng, K.; Wang, X.; Wang, S. An ABA down-regulated bHLH transcription repressor gene, bHLH129 regulates root elongation and ABA response when overexpressed in Arabidopsis. Sci. Rep. 2015, 5, 17587.
- Wang, W.; Zhu, J.; Lu, Y. Overexpression of AtbHLH112 suppresses lateral root emergence in Arabidopsis. Funct. Plant Biol. 2014, 41, 342–352.
- Lorenzo, O.; Chico, J.M.; Saénchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950.
- Anderson, J.P.; Badruzsaufari, E.; Schenk, P.M.; Manners, J.M.; Desmond, O.J.; Ehlert, C.; Maclean, D.J.; Ebert, P.R.; Kazan, K. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 2004, 16, 3460–3479.
- Yadav, V.; Mallappa, C.; Gangappa, S.N.; Bhatia, S.; Chattopadhyay, S. A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 2005, 17, 1953–1966.
- Zheng, K.; Wang, Y.; Wang, S. The non-DNA binding bHLH transcription factor Paclobutrazol Resistances are involved in the regulation of ABA and salt responses in Arabidopsis. Plant Physiol. Biochem. 2019, 139, 239–245.
- Takahashi, Y.; Ebisu, Y.; Kinoshita, T.; Doi, M.; Okuma, E.; Murata, Y.; Shimazaki, K. bHLH transcription factors that facilitate K+ uptake during stomatal opening are repressed by abscisic acid through phosphorylation. Sci. Signal. 2013, 6, ra48.
- Takahashi, Y.; Ebisu, Y.; Shimazaki, K. Reconstitution of abscisic acid signaling from the receptor to DNA via bHLH transcription factors. Plant Physiol. 2017, 174, 815–822.
- Moreno, J.E.; Moreno-Piovano, G.; Chan, R.L. The antagonistic basic helix-loop-helix partners BEE and IBH1 contribute to control plant tolerance to abiotic stress. Plant Sci. 2018, 271, 143–150.
- Jiang, Y.; Yang, B.; Deyholos, M.K. Functional characterization of the Arabidopsis bHLH92 transcription factor in abiotic stress. Mol. Genet. Genomics. 2009, 282, 503–516.
- Bai, M.Y.; Fan, M.; Oh, E.; Wang, Z.Y. A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 2012, 24, 4917–4929.
- Oh, E.; Zhu, J.; Bai, M.Y.; Arenhart, R.A.; Sun, Y.; Wang, Z.Y. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 2014, 3, e03031.
- Zhang, L.; Bai, M.Y.; Wu, J.; Zhu, J.; Wang, H.; Zhang, Z.; Wang, W.; Sun, Y.; Zhao, J.; Sun, X.; et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 2009, 21, 3767–3780.
- Hyun, Y.; Lee, I. KIDARI, encoding a non-DNA binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol. Biol. 2006, 61, 283–296.
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. ATBS1 INTERACTING FACTORs negatively regulate Arabidopsis cell elongation in the triantagonistic bHLH system. Plant Signal. Behav. 2013, 8, e23448.
- Wang, F.; Gao, Y.; Liu, Y.; Zhang, X.; Gu, X.; Ma, D.; Zhao, Z.; Yuan, Z.; Xue, H.; Liu, H. BES1-regulated BEE1 controls photoperiodic flowering downstream of blue light signaling pathway in Arabidopsis. N. Phytol. 2019, 223, 1407–1419.
- Malinovsky, F.G.; Batoux, M.; Schwessinger, B.; Youn, J.H.; Stransfeld, L.; Win, J.; Kim, S.K.; Zipfel, C. Antagonistic regulation of growth and immunity by the arabidopsis basic helix-loop-helix transcription factor HOMOLOG OF BRASSINOSTEROID ENHANCED EXPRESSION2 INTERACTING WITH INCREASED LEAF INCLINATION1 BINDING bHLH1. Plant Physiol. 2014, 164, 1443–1455.
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 2005, 120, 249–259.
- Cifuentes-Esquivel, N.; Bou-Torrent, J.; Galstyan, A.; Gallemí, M.; Sessa, G.; Salla Martret, M.; Roig-Villanova, I.; Ruberti, I.; Martínez-García, J.F. The bHLH proteins BEE and BIM positively modulate the shade avoidance syndrome in Arabidopsis seedlings. Plant J. 2013, 75, 989–1002.
- Li, X.; Duan, X.; Jiang, H.; Sun, Y.; Tang, Y.; Yuan, Z.; Guo, J.; Liang, W.; Chen, L.; Yin, J.; et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006, 141, 1167–1184.
- Jeong, J.; Kim, K.; Kim, M.E.; Kim, H.G.; Heo, G.S.; Park, O.K.; Park, Y.-I.I.; Choi, G.; Oh, E. Phytochrome and ethylene signaling integration in Arabidopsis occurs via the transcriptional regulation of genes co-targeted by PIFs and EIN3. Front. Plant Sci. 2016, 7, 1055.




